Abstract
The marine diatom Phaeodactylum tricornutum is attracting considerable interest as a candidate for biofuel production due to its fast growth and high lipid content. Nitrogen deficiency can increase the lipid content in certain microalgae species, including P. tricornutum. However, the molecular basis of such changes remains unclear without analyzing metabolism at the proteomic level. We attempted to systematically analyze protein expression level changes of P. tricornutum upon N deprivation. We observed translational level changes that could overall redirect the metabolic network from carbon flux towards lipid accumulation. N deprivation led to an increase in the expression of genes involved in nitrogen assimilation and fatty acid biosynthesis and a concomitant decrease in photosynthesis and lipid catabolism enzymes. These molecular level changes are consistent with the observed physiological changes, e.g., in photosynthesis rate and saturated lipid content. Our results provide information at the proteomic level of the key enzymes involved in carbon flux towards lipid accumulation in P. tricornutum and suggest candidates for genetic manipulation in microalgae breeding for biodiesel production.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The development of microalgal biodiesel is a major focus as an environmentally and economically feasible alternative to fossil fuels. Marine diatoms provide most of the organic matter as food for sea life. Of them, Phaeodactylum tricornutum, widely used as live feed for fishery, has a lipid content of up to 20–60 % (cell dry weight) under controlled growth conditions. Thus, it is attracting increasing interest as a candidate for biodiesel production.
Nitrogen (N) is one of the major constituents of many cellular compounds such as proteins, nucleic acids, and lipids. It is known that N deficiency can reduce primary carbon metabolism and increase lipid content in certain microalgae species. Intensive research efforts have been devoted to examining the effects of nitrogen starvation on the growth rate, lipid content, cell density, cell size, and pigment composition of microalgae. Microalgae species that have a high lipid content under nitrogen starvation conditions, include P. tricornutum (Qian and Michael 1993), and the green algae Chlorella spp. (Illman et al. 2000), Botryococcus braunii (Zhila et al. 2005), and Chlamydomonas reinhardtii (Miller et al. 2010), whereas others, such as Dunaliella salina, do not (Ben-Amotz et al. 1985). Such accumulation of lipids under nitrogen starvation can also be affected by other environmental conditions. For example, in the marine microalga Nannochloropsis, it was found that combined high salinity and light intensity impairs lipid productivity under nitrogen starvation (Pal et al. 2011). With the increased availability of sequenced genomes, fatty acid profiling has been carried out in C. reinhardtii (James et al. 2011), and changes in transcript abundance have been used to predict the diversion of metabolism following nitrogen deprivation (Miller et al. 2010). Recently, transcriptional analysis on P. tricornutum has extensively characterized global gene expression during enhanced lipid production as a consequence of nitrogen depletion (Valenzuela et al. 2012). The proteome of another model diatom, Thalassiosira pseudonana, at the onset of nitrogen starvation has also recently been compared with that of nitrogen-replete cells by two-dimensional gel electrophoresis (2-DE), revealing differences in the responses of central carbon metabolism under nitrogen starvation between diatoms, green algae, and higher plants (Hockin et al. 2011).
So far, no proteomic studies have been carried out in diatoms to systematically elucidate the molecular shifts towards neutral lipid accumulation at the proteomic level. In this study, we acquired proteomic data in P. tricornutum following N deprivation (−N) to illustrate the multiple levels of regulation redirecting metabolism towards lipid accumulation.
Materials and methods
Phaeodactylum tricornutum was obtained from the Algal Collection Center of Jinan University (no. FACHB-863). Cells were grown as batch cultures in flasks containing f/2-Si medium (omitting Na2SiO3·9H2O). Cultures were routinely cultivated in an artificial climate incubator under constant irradiance (200 μmol photons m−2 s−1), temperature (21 ± 0.5 °C), and 12/12-h light/dark cycles.
Algal culture (1.5 L) in the late logarithmic growth phase was centrifuged (4,400 rpm, 10 min, 4 °C) and the pelleted cells were washed twice with N-free f/2-Si medium. Next, cells were collected by centrifugation (4,400 rpm, 15 min), and pelleted cells were reinoculated into 1.5 L of N-free f/2-Si medium. Half of the cultures were supplemented with N. The resuspended cultures were evenly divided into 12 250-mL flasks and cultures supplemented with N as control. Cells were cultured for a further 2 days for fatty acid profiling or 2D analysis.
Protein extraction and 2-DE
Pelleted diatom cells were rinsed twice with sterile water and ground with liquid nitrogen into a fine powder. The powder was transferred into 1.5-mL tubes at 4 °C, and 500 μL of lysis buffer was added. Cells were lysed for 30 min. Cellular debris was removed by centrifugation (15,000×g, 30 min, 4 °C), prechilled acetone (five times supernatant volume) was added to precipitate the protein, and the solution was kept at −20 °C for 1 h. Crude protein was collected by centrifugation as previously described. The precipitate was washed three times with ice-cold acetone and recovered by centrifugation as previously described. Residual acetone was volatilized under a sterile, cool, ventilated environment for 10 min. Finally, the protein pellet was resuspended in 200 μL rehydration buffer and mixed by vigorous agitation for 30 min at 4 °C to solubilize the protein completely. The insoluble material was removed by centrifugation as previously described. Protein concentration was determined by a modified Bradford assay (Bio-Rad, USA).
2-DE was performed with an Ettan IPGphor III Isoelectric Focusing System (GE, USA). A 250-μL solution containing 200 μg total protein was used for passive rehydration in 13 cm immobilized pH gradient strips (Ready Strip IPG strips pH3-10 NL, GE, USA).
Image analysis and protein identification
Images of silver-stained gels acquired with an image scanner (Amersham Biosciences, USA) were analyzed using ImageMaster2D Platinum 6.0 (GE, USA). Spots with statistical significance (Student’s t test, p < 0.05) were considered, and spots with an abundance ratio of at least two were selected as differentially expressed proteins. Differentially expressed spots were manually excised from the gel. In-gel digestion and mass spectrometry analysis were performed according to Zhao et al. (2005). The identified proteins were then matched to specific processes or functions by querying Gene Ontology (http://www.geneontology.org/).
Analysis of photosynthetic efficiency
Chlorophyll fluorescence is an ideal probe for the characterization of plant photosynthesis. The chlorophyll fluorescence in P. tricornutum culture was measured with a Handy PEA chlorophyll fluorometer (Hansatech Instruments Ltd., UK). Algae culture was kept in the dark for 20 min prior to exposure to saturation pulse light (3,000 mol photons m−2 s−1) for 1 s, followed by measurement of F v/F m (the ratio of variable/maximum fluorescence). The chlorophyll fluorescence parameter F v/F m is an empirically verifiable index of photosynthetic performance and acclimation status (Marillia et al. 2003). It corresponds to the maximum photochemical quantum yield of photosystem II (PSII) reaction center and reflects the light energy conversion efficiency of photosynthesis.
Analysis of fatty acids by GC-MS
A 250-mL aliquot of diatom culture was harvested 48 h after N deprivation (−N). Lipids were extracted according to Lepage, with minor modifications (Liu and Curtiss 2009). Fatty acid composition was determined by gas chromatography–mass spectrometry (GC-MS) at the Institute of Microbiology of Guangdong, China. A DB-5 quartz capillary column with specifications 30 m × 0.25 mm × 0.25 μm was used. The column temperature was programmed from 60 to 160 °C at a rate of 10 °C min−1 and to a final temperature of 250 °C at a rate of 2.5 °C min−1. Injector was set at 280 °C and splitless was 1 μl. Mass spectrum transmission line was set at 200 °C. Fatty acid composition was identified with the equipped NBS spectrum library. Peak areas were calculated by normalization to acquire the percent contents.
Results
Differential protein expression in P. tricornutum under −N
To quantify the protein-level responses of P. tricornutum to −N, 2-DE was performed on equal amounts of cell protein extracts from N-deprived and control groups. Within a pair of N-deprived and control cells, more than 1,200 spots were automatically matched between the 2-DE gels by Image Master Platinum. Forty-two significantly differentially expressed spots with a volume ratio >2.0 (p < 0.05) were successfully identified (Fig. 1), including 37 nonredundant proteins. Among them, 15 were upregulated and 22 were downregulated. Detailed information is listed in Table 1.
Representative 2-DE gels of proteins of N-deprived (−N) and N-replete (control) cultures. Left control, right −N. Molecular weight and pH are labeled at the side and top. Spot indices correspond to the IDs in Table 1. Differentially regulated proteins are indicated by arrows
Mapping protein expression data to KEGG pathways
To study the changes of metabolic enzymes in response to −N, all proteins identified in the 2D proteomic analysis were mapped to the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway IDs using both the KEGG database and homology mapping. Direct neighbors and second degree interactions that could connect two identified proteins were included to construct the metabolic connections shown in Fig. 2. Expression level changes were represented by color intensity. Detailed pathway involvement of diverse enzymes that redirect carbon flux to lipid accumulation is discussed in the following sections.
Pathway participation of significantly changed proteins. Proteins identified were mapped to KEGG pathways, and those involved in the pathways were assigned KEGG IDs. Each node represents a single protein indicated by PHATRDRAFT ID, and different colors represent the differential expression changes at the protein level. All first degree interactions and second degree interactions that allow the connection of two identified proteins are included in the figure
Proteins associated with N assimilation increased
Proteins identified were mapped to KEGG pathways as depicted in Fig. 2. Firstly, the primary response of the cells under −N was considered to be the upregulation of nitrogen assimilation enzymes, which assists diatom cells to efficiently utilize the trace amount of nitrogen in the culture. Consistently, we observed a large increase in the abundance of glutamine synthase (GS, PHATRDRAFT_22357; Fig. 2), the key enzyme involved in ammonia assimilation in both plants and Gram-negative microorganisms (Stacey et al. 1979).
Proteins responsible for amino acid metabolism
Most of the proteins associated with amino acid metabolism showed decreased expression after −N (Table 1). Ornithine aminotransferase (PHATRDRAFT_27726) is a mitochondrial enzyme containing pyridoxal-5′-phosphate as a cofactor which catalyzes the interconversion between ornithine and glutamate semialdehyde (Delauney et al. 1993). Its expression level was decreased under −N (Fig. 2). A significant decrease was observed for protein PHATRDRAFT_33928, which encodes a putative 3-isopropylmalate dehydrogenase. This enzyme is involved in the leucine biosynthesis pathway, which catalyzes the dehydrogenation and concomitant decarboxylation of the 3-isopropylmalate substrate. Thus, biosynthesis of amino acids decreased following −N, as further indicated by the determined 25 % decrease in protein content per cell following −N (Fig. 3a). A decrease was observed for a putative serine/threonine protein kinase (PHATRDRAFT_44441; Fig. 2). The kinases of this family have been suggested to be involved in apoptosis (Cross et al. 2000). One exception to the overall decreased expression was cysteine synthase (CysK; PHATRDRAFT_bd542). CysK is responsible for the formation of cysteine from O-acetylserine and hydrogen sulfide with concomitant release of acetic acid.
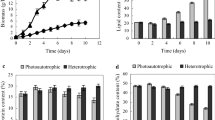
Protein content and photosynthetic activity under −N. a Protein content. Total proteins were extracted from control and N-deprived diatom cells at 2 days after medium substitution. Protein concentration was determined by the Bradford assay. b Photosynthetic activity. The chlorophyll fluorescence parameter F v/F m was monitored for algal cultures, and standard bars represent the standard deviation of samples in triplicate
Decreased expression of the light-harvesting complex involved in photosynthesis following −N
Photosynthetic carbon fixation requires nitrogen for producing proteins that participate in electron transport and catalyze photosynthetic reactions. A lower protein content caused by nitrogen starvation could limit electron transport (Hockin et al. 2011). Consistently, a noticeable downregulation of expression was observed in a cluster of proteins involved in light harvesting in photosynthesis. In total, six markedly altered spots were identified as members of fucoxanthin chlorophyll a/c proteins (FCP). FCPs are homologs of the light-harvesting complexes of higher plants, which perform a similar function in photosynthesis in brown algae and diatoms. An FCP complex, FCPb, has been described for the diatom Cyclotella meneghiniana, and an association between FCPb and photosystem I (PSI) has been proposed (Veith et al. 2009). Among the six spots identified, only one showed an increase in expression (PHATRDRAFT_44601). The rest showed decreased expressions at the protein level. Interestingly, three spots were identified as the same protein (PHATRDRAFT_54065; Fig. 2), and they exhibited the same size of 22 kDa but with a series of adjacent pIs, which is characteristic of proteins with multiple phosphorylation sites. Another enzyme, geranylgeranyl reductase (PHATRDRAFT_50650), which participates in carotenoid biosynthesis, also exhibited a large decrease in expression under −N. This effect is consistent with that of the fucoxanthin chlorophyll a/c light-harvesting protein.
Next, we measured photosynthesis activity to provide physiological evidence for the proteomics data. Since chlorophyll fluorescence is an ideal probe for characterizing photosynthesis (Marillia et al. 2003), chlorophyll fluorescence in a P. tricornutum culture was measured. As shown in Fig. 3b, the maximum energy conversion efficiency indicated by F v/F m in the algal culture following −N decreased by 7 %. This result further indicates that the light energy absorption and photosynthetic conversion efficiency decreased following −N.
Carbon and energy metabolism and carbon flux towards lipid accumulation
The majority of identified proteins (17 in total) were categorized as being involved in biological process of carbon and energy metabolism. Plastidic ribulose-phosphate 3-epimerase (PHATRDRAFT_53935), which catalyzes the reversible epimerization of d-ribulose 5-phosphate to d-xylulose 5-phosphate, showed a large increase in expression under −N. The enzyme is located in the thylakoid membrane and participates in several important metabolic pathways, including carbohydrate biosynthesis and the Calvin cycle. Increased expression of this enzyme may contribute to the increased yield of RuBP, thus promoting carbon fixation and the production of 3-P-glyceraldehyde, which is the raw material for triglyceride biosynthesis, or it could be used in glucosamine synthesis (Nowitzki et al. 1995). Expression of another plastidic enzyme, plastidic adenosine triphosphate (ATP) synthase subunit beta (PhtrCp050), also increased following −N. ATP synthase is a large mushroom-shaped complex consisting of a stalk (F0 portion) spanning the thylakoid membrane and a cap (F1 portion) on the stromal side. Protons are pumped by PSII and PSI into the thylakoid lumen and move across the thylakoid membrane for ATP synthesis on the stromal side and, thus, are available for use in CO2 fixation occurring in the stroma. Increased production of plastidic ATP synthase could, therefore, promote carbon fixation under −N. Moreover, another enzyme involved in CO2 acquisition, a putative intracellular beta-type carbonic anhydrase (CA; PHATRDRAFT_45443; Fig. 2), showed considerably reduced expression under −N. CA catalyzes the reversible dehydration of HCO3 − to CO2 and plays important roles in various biological processes, such as ion exchange, respiration, pH homeostasis, CO2 acquisition, and photosynthesis. Consistent with our observation, CA has also been detected at reduced levels in high CO2-grown P. tricornutum (Santos et al. 2011).
In total, four glyceraldehyde 3-phosphate dehydrogenases (GAPDH; PHATRDRAFT_22122, PHATRDRAFT_32747, PHATRDRAFT_25308, and PHATRDRAFT_50738; Fig. 2) were identified, and all showed decreased expression following −N. GAPDH is involved in gluconeogenesis, glycolysis, the phosphopentose shunt, and fatty acid biosynthesis by catalyzing the reversible oxidative phosphorylation of d-glyceraldehyde 3-phosphate in the presence of NAD and inorganic phosphate, thereby controlling an important energy-yielding step in glycolysis in carbohydrate metabolism (Blanc et al. 2010). It is also noteworthy that the abundance of transaldolase (Tal, PHATRDRAFT_28222; Fig. 2) showed a substantial increase in −N. This nearly ubiquitous enzyme is involved in central carbon metabolism, i.e., the non-oxidative pentose phosphate pathway, transferring a dihydroxyacetone group from donor compounds (fructose 6-phosphate or sedoheptulose 7-phosphate) to aldehyde acceptor compounds (Samland et al. 2012). The pentose phosphate pathway produces a large amount of NADPH by complete oxidation of glucose 3-phosphate, which provides the main reducing power of cells necessary for various synthetic reactions, such as fatty acid synthesis and ammonia assimilation.
Phosphomannomutase (PMM; PHATRDRAFT_28882; Fig. 2), which catalyzes the conversion of mannose-6-phosphate to mannose-1-phosphate as an essential step in mannose activation and the biosynthesis of glycoconjugates in all eukaryotes, showed a marked decrease. PMM is one of three enzymes that function in the biosynthesis of guanosine diphosphate (GDP)-mannose from the glycolytic intermediate fructose 6-phosphate. PMM from the red alga Galdieria sulphuraria has been shown to be bifunctional, utilizing both glucose phosphates and mannose phosphates as substrates (Nguyen et al. 2011). Decreased expression of PMM following −N would thus affect GDP-mannose biosynthesis and redirect carbon flux back to lipid accumulation. Phosphoglycerate mutase participates in glycolysis and is mainly responsible for the transfer of a phosphate group. A putative PGAM_5 (PHATRDRAFT_26201) showed a large increase in expression under −N, accelerating glycolysis and supplying the raw material for lipid biosynthesis.
Pyruvate dehydrogenase complex (PDC) is an important enzyme in pyruvate metabolism. Pyruvate dehydrogenase E1 component (PHATRDRAFT_20360), which catalyzes the first step of PDC, showed increased expression under −N, thereby accelerating the conversion of pyruvate to acetyl-CoA.
Alterations of proteins involved in lipid metabolism
Among the identified proteins associated with lipid biosynthesis, a notable upregulation of enoyl-acyl carrier protein (ACP) reductase (ENR; PHATRDRAFT_10068) was detected. ENR is one of the major components of the fatty acid synthase complex, which catalyzes the reduction of the trans-2,3 double bond of a saturated acyl chain in the fatty acid elongation cycle. During seed development, ENR enzyme activity in Brassica napus has been shown to increase during the period of storage lipid accumulation (Kater et al. 1991). In the present study, N deprivation had a large effect on the fatty acid composition of P. tricornutum cells determined by GC-MS as shown in Fig. 4. The unsaturated fraction of lipids in P. tricornutum decreased by 16 % following −N. The fraction of polyunsaturated fatty acids, including C20:5 and C22:6, showed a dramatic decrease of around 40 %, which is consistent with the elevated expression of ENR.
Additionally, the proportion of saturated fatty acids and monounsaturated fatty acids showed an increase of 45 and 61 %, respectively. According to the analysis of fatty acid composition, the saturation of fatty acids increased in P. tricornutum following −N.
Proteins involved in genetic information processing
Heat shock proteins (HSPs) are a group of functionally related proteins responsible for protein folding and unfolding. High-level expression of HSPs can be triggered by exposure to different environmental stress conditions, including exposure of the cell to nitrogen deficiency (Choi et al. 2007). We identified similar trends in N-deprived cells, including two HSPs, which were dramatically upregulated: Hsp70 (PHATRDRAFT_41417) and Hsp90 (PHATRDRAFT_55215) (Fig. 2).
A putative elongation factor G (EF-G; PHATRDRAFT_11189; Fig. 2) showed higher expression following −N. The primary function of EF-G is to catalyze translocation in the elongation cycle of protein synthesis. In contrast, a slight decrease was observed for another protein involved in genetic information processing, the polyadenylate binding protein (PHATRDRAFT_23959; Fig. 2), which binds mRNA and participates in mRNA polyadenylation by interacting selectively and noncovalently with an RNA molecule (Anderson et al. 1993).
Other proteins involved in various cellular functions
In total, 10 other significantly changed proteins, which could not be categorized as being associated with particular biological processes, were grouped into the miscellaneous category (Table 1). Two peroxidases were downregulated under −N, including a putative ascorbate peroxidase (PHATRDRAFT_47395) and cytochrome c peroxidase (PHATRDRAFT_13244). Further, superoxide dismutase [Fe/Mn] (PHATRDRAFT_12583), mainly responsible for the superoxide metabolic process and for destroying radicals, showed a large decrease.
An NmrA-like family protein (PHATRDRAFT_46721), which is a redox sensor protein, became abundant following −N. This protein has been shown to be a negative transcriptional regulator in Aspergillus nidulans responsible for modulating the activity of a transcription factor AreA (Shi and Shi 2004). AreA is a major gene involved in the repression of nitrogen metabolites in A. nidulans (Wilson and Arst 1998). NmrA presents an NAD+-binding domain for protein function, thus defining a class of NAD(P)+-dependent proteins involved in sensing metabolic parameters (Lamb et al. 2004; Shi and Shi 2004).
Discussion
Expression level changes of proteins following N deprivation in the diatom P. tricornutum were investigated in this study. Overall pathway involvement of diverse enzymes redirected carbon flux to lipid accumulation. Above all, the upregulation of nitrogen assimilation enzymes such as GS assisted diatom cells to efficiently utilize the trace amount of nitrogen in the culture.
Decreased expression of most of the proteins associated with amino acid metabolism indicated that amino acid biosynthesis was at least partially inhibited due to the absence of nitrogen. These findings may implicate that the biosynthesis of amino acids decreased following −N, reserving metabolic substrates for lipid production. The family of serine/threonine protein kinase is involved in apoptosis (Cross et al. 2000), thus its reduction could repress apoptosis to maintain cell growth. One exception to the overall decreased expression was CysK. In bacteria, CysK differs from CysM in that it can also use sulfide in place of thiosulfate, producing cysteine instead of cysteine thiosulfonate. It has been shown in plants that the overexpression of CysK could alleviate reactive oxygen species (ROS) damage (Xu et al. 2011). ROS are considered to be the major source of damage to cells under biotic and abiotic stresses in plants (Candan and Tarhan 2003). Therefore, the increased expression of CysK could be a stress response to −N.
The light-harvesting complex involved in photosynthesis, including six members of FCP, showed decreased expression. This result was consistent with findings for the diatom Cyclotella cryptica, where in vivo experiments using antibodies against phosphothreonine residues and in vitro studies using [γ-32P]ATP showed that fucoxanthin chlorophyll a/c binding proteins of 22 kDa became phosphorylated (Brakemann et al. 2006). This suggests that −N could decelerate the light-harvesting process in photosynthesis. The maximum energy conversion efficiency decreased by 7 % following −N, further indicating the decrease in light energy absorption and photosynthetic conversion efficiency. Downregulation of the light-harvesting complex under −N effectively reduces light energy absorption. Furthermore, the decrease in photosynthetic activity inhibits the relevant carbon fixation pathway. Therefore, alternative metabolic pathways may occur, which compensate for carbon assimilation and eventually lead to high lipid accumulation under −N. Taken together, the decreased expression of proteins involved in photosynthesis and the inhibition of photosynthetic activity indicate that the observed high lipid accumulation can only be partially attributed to metabolic substrates from photosynthesis but is mainly due to the conversion of other intracellular substrates.
Carbon and energy metabolism and carbon flux were redirected towards lipid accumulation, as implicated by the observation that the majority of identified proteins (17 in total) were categorized as being involved in the biological process of carbon and energy metabolism. Carbon fixation was promoted, as revealed under −N. The decreased expression of GAPDH in combination with the increased expression of Tal could consequently promote the production of NADPH and eventually contribute to the direction of carbon flux towards lipid accumulation under −N. Decreased expression of PMM would affect GDP-mannose biosynthesis and also redirect carbon flux back to lipid accumulation. Increased expression of pyruvate dehydrogenase E1 component, which catalyzes the first step of PDC, thereby accelerated the conversion of pyruvate to acetyl-CoA. This could provide the substrate for the TCA cycle and also the raw material for fatty acid biosynthesis following the conversion of acetyl-CoA to citrate. Among the identified proteins involved in lipid biosynthesis, the notable upregulation of ENR was in accordance with the increased expression in B. napus during the period of storage lipid accumulation (Kater et al. 1991).
Increased expression of proteins involved in genetic information processing such as HSPs responsible for protein folding and unfolding was triggered by exposure of the cell to nitrogen deficiency. N starvation is known to cause oxidative stress in plants, which can be reflected as changes in a series of stress responsive proteins. In diatoms, it has been found that a lower protein content following nitrogen starvation could limit electron flow through the photosynthetic apparatus, resulting in elevated production of ROS, and thus oxidative stress (Hockin et al. 2011). The slight decrease of the polyadenylate binding protein, which binds mRNA and participates in mRNA polyadenylation (Anderson et al. 1993), indicates that mRNA maturation was affected under −N, implying that protein translation was reduced and cells reserved the metabolic intermediates, possibly for lipid synthesis.
There were still 10 other significantly changed proteins involved in various cellular functions. For instance, two peroxidases and superoxide dismutase [Fe/Mn] were downregulated, largely reflecting the elevated oxidative stress following −N. The high abundance of NmrA, which presents an NAD+-binding domain for protein function (Lamb et al. 2004; Shi and Shi 2004), suggesting that the level of NADPH significantly increased under −N to meet the demands of fatty acid biosynthesis.
In conclusion, the dataset presented should be an important contribution to the community since many studies are interested in the process leading to oil accumulation in microalgae. Analysis of carbon flux, metabolome, and TAG accumulation will be needed to complement comparative proteomic analyses, so as to assess flux through the TAG accumulation relevant metabolic network at a systems biology level.
References
Anderson J, Wilson S, Datar K, Swanson M (1993) NAB2: a yeast nuclear polyadenylated RNA-binding protein essential for cell viability. Mol Cell Biol 13:2730–2741
Ben-Amotz A, Tornabene TG, Thomas WH (1985) Chemical profile of selected species of microalgae with emphasis on lipids. J Phycol 21:72–81
Blanc G, Duncan G, Agarkova I, Borodovsky M, Gurnon J, Kuo A, Lindquist E, Lucas S, Pangilinan J, Polle J, Salamov A, Terry A, Yamada T, Dunigan DD, Grigoriev IV, Claverie JM, Van Etten JL (2010) The Chlorella variabilis NC64A genome reveals adaptation to photosymbiosis, coevolution with viruses, and cryptic sex. Plant Cell 22:2943–2955
Brakemann T, Schlörmann W, Marquardt J, Nolte M, Rhiel E (2006) Association of fucoxanthin chlorophyll a/c-binding polypeptides with photosystems and phosphorylation in the centric diatom Cyclotella cryptica. Protist 157:463–475
Candan N, Tarhan L (2003) The correlation between antioxidant enzyme activities and lipid peroxidation levels in Mentha pulegium organs grown in Ca2+, Mg2+, Cu2+, Zn2+ and Mn2+ stress conditions. Plant Sci 165:769–776
Choi Y, Kwon K, Lee J, Woo S (2007) Expression of the rice cytoplasmic cysteine synthase gene in tobacco reduces ozone-induced damage. Plant Biotechnol Rep 1:93–100
Cross T, Scheel-Toellner D, Henriquez N, Deacon E, Salmon M, Lord J (2000) Serine/threonine protein kinases and apoptosis. Exp Cell Res 256:34–41
Delauney A, Hu C, Kishor PK, Verma D (1993) Cloning of ornithine δ-aminotransferase cDNA from Vigna aconitifolia by trans-complementation in Escherichia coli and regulation of proline biosynthesis. J Biol Chem 25:18673–18678
Hockin NL, Mock T, Mulholland F, Kopriva S, Malin G (2011) The response of diatom central carbon metabolism to nitrogen starvation is different from that of green algae and higher plants. Plant Physiol 158:299–312
Illman AM, Scragg AH, Shales SW (2000) Increase in Chlorella strains calorific values when grown in low nitrogen medium. Enzyme Microb Technol 27:631–635
James GO, Hocart CH, Hillier W, Chen H, Kordbacheh F, Price GD, Djordjevic MA (2011) Fatty acid profiling of Chlamydomonas reinhardtii under nitrogen deprivation. Bioresource Technol 102:3343–3351
Kater MM, Koningstein GM, Nijkamp HJJ, Stuitje AR (1991) cDNA cloning and expression of Brassica napus enoyl-acyl carrier protein reductase in Escherichia coli. Plant Mol Biol 17:895–909
Lamb HK, Ren J, Park A, Johnson C, Leslie K, Cocklin S, Thompson P, Mee C, Cooper A, Stammers DK, Hawkins AR (2004) Modulation of the ligand binding properties of the transcription repressor NmrA by GATA-containing DNA and site-directed mutagenesis. Protein Sci 13:3127–3138
Liu X, Curtiss R (2009) Nickel-inducible lysis system in Synechocystis sp. PCC 6803. Proc Natl Acad Sci 106:21550–21554
Marillia E-F, Micallef BJ, Micallef M, Weninger A, Pedersen KK, Zou J, Taylor DC (2003) Biochemical and physiological studies of Arabidopsis thaliana transgenic lines with repressed expression of the mitochondrial pyruvate dehydrogenase kinase. J Exp Bot 54:259–270
Miller R, Wu G, Deshpande RR, Vieler A, Gartner K, Li X, Moellering ER, Zauner S, Cornish AJ, Liu B, Bullard B, Sears BB, Kuo MH, Hegg EL, Shachar-Hill Y, Shiu SH, Benning C (2010) Changes in transcript abundance in Chlamydomonas reinhardtii following nitrogen deprivation predict diversion of metabolism. Plant Physiol 154:1737–1752
Nguyen HM, Baudet M, Cuiné S, Adriano J-M, Barthe D, Billon E, Bruley C, Beisson F, Peltier G, Ferro M, Li-Beisson Y (2011) Proteomic profiling of oil bodies isolated from the unicellular green microalga Chlamydomonas reinhardtii: with focus on proteins involved in lipid metabolism. Proteomics 11:4266–4273
Nowitzki U, Wyrich R, Westhoff P, Henze K, Schnarrenberger C, Martin W (1995) Cloning of the amphibolic Calvin cycle/OPPP enzyme d-ribulose-5-phosphate 3-epimerase (EC 5.1.3.1) from spinach chloroplasts: functional and evolutionary aspects. Plant Mol Biol 29:1279–1291
Pal D, Khozin-Goldberg I, Cohen Z, Boussiba S (2011) The effect of light, salinity, and nitrogen availability on lipid production by Nannochloropsis sp. Appl Microb Biotechnol 90:1429–1441
Qian K, Michael AB (1993) Light and nitrogen deficiency effects on the growth and composition of Phaeodactylum tricornutum. Appl Biochem Biotechnol 38:93–103
Samland AK, Baier S, Schürmann M, Inoue T, Huf S, Schneider G, Sprenger GA, Sandalova T (2012) Conservation of structure and mechanism within the transaldolase enzyme family. FEBS J 279:766–778
Santos CNS, Koffas M, Stephanopoulos G (2011) Optimization of a heterologous pathway for the production of flavonoids from glucose. Metab Eng 13:392–400
Shi Y, Shi Y (2004) Metabolic enzymes and coenzymes in transcription—a direct link between metabolism and transcription? Trend Genet 20:445–452
Stacey G, Van Baalen C, Tabita FR (1979) Nitrogen and ammonia assimilation in the cyanobacteria: regulation of glutamine synthetase. Arch Biochem Biophy 194:457–467
Valenzuela J, Mazurie A, Carlson RP, Gerlach R, Cooksey KE, Peyton BM, Fields MW (2012) Potential role of multiple carbon fixation pathways during lipid accumulation in Phaeodactylum tricornutum. Biotechnol Biofuels 5/1:40
Veith T, Brauns J, Weisheit W, Mittag M, Büchel C (2009) Identification of a specific fucoxanthin-chlorophyll protein in the light harvesting complex of photosystem I in the diatom Cyclotella meneghiniana. BBA - Bioenergetics 1787:905–912
Wilson RA, Arst HN Jr (1998) Mutational analysis of AREA, a transcriptional activator mediating nitrogen metabolite repression in Aspergillus nidulans and a member of the “streetwise” GATA family of transcription factors. Microbiol Mol Biol Rev 62:586–596
Xu C, Jiang Z, Huang B (2011) Nitrogen deficiency-induced protein changes in immature and mature leaves of creeping bentgrass. J Am Soc Hortic Sci 136:399–407
Zhao C, Wang J, Cao M, Zhao K, Shao J, Lei T, Yin J, Hill G, Xu N, Liu S (2005) Proteomic changes in rice leaves during development of field-grown rice plants. Proteomics 5:961–972
Zhila NO, Kalacheva GS, Volova TG (2005) Influence of nitrogen deficiency on biochemical composition of the green alga Botryococcus. J Appl Phycol 17:309–315
Acknowledgments
This work was supported by the National Technologies Program (2011BAD14B03), the Science and Technology Project of Guangdong, and the Fundamental Research Funds for the Central Universities.
Author information
Authors and Affiliations
Corresponding author
Additional information
Zhi-Kai Yang and Yu-Han Ma equally contributed to this work.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Yang, ZK., Ma, YH., Zheng, JW. et al. Proteomics to reveal metabolic network shifts towards lipid accumulation following nitrogen deprivation in the diatom Phaeodactylum tricornutum . J Appl Phycol 26, 73–82 (2014). https://doi.org/10.1007/s10811-013-0050-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10811-013-0050-3