Abstract
Purpose
This paper aimed to assess the diagnostic utility of a newly developed gene-based technology-nanopore targeted sequencing (NTS) in suspected endophthalmitis patients.
Methods
This retrospective study included 43 patients (44 eyes) with suspected endophthalmitis. NTS was applied along with microbiological culture to detect unknown pathogens in intraocular fluid samples. The diagnostic utility of NTS was mainly evaluated from three aspects, including the positivity rate of bacterial/fungal presence, diagnostic turnaround time and the frequency of change in treatment based on etiology test results. Non-parametric, two-sided Wilcoxon rank sum test, the McNemar’s test and the kappa statistic were used for statistical comparisons.
Results
NTS showed significant advantages over traditional culture in positivity rates and diagnostic time (P < 0.001, kappa = 0.082; Z = −5.805, P < 0. 001). As regards antibiotic strategy, 17 patients (39.53%) and 5 patients (11.63%) underwent medication change following NTS and culture results respectively (P < 0.001, kappa = 0.335). With reasonable use of antibiotic and surgical intervention, most patients responded favorably, judged by significantly improved visual acuity (Z = −4.249, P < 0.001). The mean duration of hospitalization was 8.49 ± 2.45 days (range, 1–16 days).
Conclusion
The high efficiency feature of NTS in pathogen detection renders it a valuable supplementary to traditional culture. Additionally, it has facilitated patients’ management for the early and precise diagnosis of endophthalmitis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Endophthalmitis is one of the most devastating ocular diseases, and it can lead to permanent vision loss within a few days. Most cases of exogenous endophthalmitis are caused by infectious pathogens introduced to the eye from either the ocular surface or the environment (such as keratitis or trauma). Endogenous endophthalmitis (EE) cases are rare but highly destructive, and it usually result from chronic systemic infections (such as liver abscesses, endocarditis, or urinary tract infections [1]). Due to the delicate anatomy of the eye and the limited ocular sample size, it has presented a diagnostic challenge to identify the diversified causative organisms in endophthalmitis cases. Prognosis may be poor owing to delayed diagnosis and lack of targeted anti-microbial treatment. Therefore, the most critical issue in diagnosing endophthalmitis is the identification of clinical microbiology, as early identification of causative pathogen(s) is imperative to guide further anti-microbial treatment.
Although the positivity rate is only about 38%-64%, culture performed in vitreous humor (VH) or aqueous humor (AH) samples remains the gold standard for the pathogen detection in endophthalmitis patients. Nevertheless, the time required for different pathogen culture ranges from a few days to several weeks, often resulting in delayed diagnosis [2]. Thus, molecular diagnostic techniques emerged. Multiplex PCR testing have the potential to identify pathogens rapidly, especially in culture-negative cases. However, this method requires primers based on prior assumptions about the species and only detects a narrow array of pathogens. Next-generation sequencing (NGS) technologies have contributed to a broad detection range and it boasts a high accuracy for laboratory diagnosis of endophthalmitis. More recently, nanopore-targeted sequencing (NTS) came to light and a set of targeted microbial tags were incorporated [3]. With targeted gene amplification, the unique advantage of its long-read and real-time analysis can be achieved.
In this article, we further verified the effectiveness of NTS by focusing on clinical-oriented aspects. The purpose of our study was to assess the diagnostic utility of NTS in suspected endophthalmitis cases and explore the role of NTS intervention in the early clinical decisions.
Materials and methods
Study design and patients
By reviewing the medical records from the hospital information system (HIS) of the Ophthalmic Center, a retrospective study was performed in the endophthalmitis patients who underwent NTS and culture simultaneously between January 1, 2018, and July 24, 2022. This study was carried out following the institutional guidelines and ethical standards of the 1964 Declaration of Helsinki and was approved by the Institutional Review Board of Renmin Hospital of Wuhan University (WDRY2019-K056). All patients provided their written informed consents to participate in this study.
The criteria included: 1. Recent history of eye surgery or penetrating ocular trauma, or with other predisposing factors; 2. Typical manifestations of significant loss of vision, ocular pain, ocular redness, etc.; 3. Marked intraocular inflammation like hypopyon and vitritis on ocular examination; 4. Intraocular fluid including aqueous humor (AH) or vitreous humor (VH) was collected and sent for NTS and microbiological culture simultaneously; 5. With a minimum 3-month follow-up.
The patients’ records were reviewed including demographic characteristics, best corrected visual acuity (BCVA) at their first visit, clinical features on slit-lamp examination, disease course and treatment, etiology test results (NTS and/or culture results), the time to confirmatory diagnosis, management details, length of hospitalization and BCVA at last follow-up.
Outcome measures
The positivity rate of bacterial/fungal presence, diagnostic turnaround time, and frequency of changes in treatment protocol based on etiology test results (defined as any change directly attributable to sequencing or culture results) are used as primary outcome measures. Continuous data were presented as mean and standard deviation, whereas categorical data were presented as the number of suffered eyes and percentage. For statistical analysis, vision was reported as mean and median logMAR vision with Snellen conversion. Non-Snellen acuities were recorded in the following fashion: a visual acuity of 2/800 on Feinbloom’s low vision chart was considered equivalent to counting fingers (CF), and it was defined as 2.6 logMAR. Likewise, we used logMAR values of 2.7, 2.8, and 2.9 to represent the vision of hand movement (HM), light perception (LP), and no light perception (NLP), respectively.
Sample collection
To limit contamination, intraocular fluid (AH or VH) samples for NTS were obtained under strictly sterile conditions. AH samples were obtained through anterior chamber paracentesis, and VH samples were obtained during the biosurgery procedure. The operations were performed by the same operator, and purulent lesions and inflammatory exudate were cleared as much as possible during the operations. All clinical specimens were then sent to the clinical laboratory with specific pretreatment. The experimental procedures were performed by well-trained laboratory technicians in a qualified laboratory (Wuhan Dgensee Clinical Laboratory Co., Ltd. Wuhan 430,075, China).
Culture method
Gram stain and KOH mount were routinely performed on aqueous humor or vitreous specimens. The remaining samples were inoculated on Columbia blood AGAR basal medium (for bacteria) and Sabouraud glucose AGAR medium (for fungi) using a BACTEC 9120 culture system (BD Diagnostics, Sparks, MD). For culture-positive cases, isolated fungi and/or bacteria were identified using the Vitek 2 Compact automated identification system (bioMerieux, Marcy L 'Etoile, Huang et al. 1061 France) and MALDI Biotyper mass spectrometry (Bruker, Marcy L 'Etoile, Huang et al. Madison, WI).
Sequencing method
Preprocessing and DNA extraction
Intraocular fluid samples were centrifuged at 20,000 ×g for 10 min. The supernatant was removed, and 200 μL of the specimen was reserved for DNA extraction (Sansure DNA Extraction Kit, Changsha, China). All primers used in this study have been described in a previously published article [, 3, 4].
NTS library construction and sequencing
Amplification of the bacterial 16S rRNA gene was performed in a 20 μL reaction system with 8 μL of extracted DNA, 2 μL of barcoded primer (10 μM), and 10 μL of 2 × KOD TM PCR Master Mix (TOYOBO) using the following cycle: 98 °C for 3 min; followed by 35 cycles at 98 °C for 10 s, 55 °C for 5 s, and 68 °C for 10 s; and a final elongation step at 68 °C for 5 min.
Amplification of the fungal internal transcribed spacers 1 and 2 (ITS1/2) was performed in the same reaction system and the primer mix without the barcode was used in the PCR procedure. The PCR product was purified with 0.8 × AMpure beads (Beckman Coulter) and eluted in 10 μL Tris–EDTA (TE) buffer. Then, 5 μL of the eluate was used for PCR with 5 μL of the barcoded ITS1/2 primer set (10 μM), and 10 μL 2 × Phusion U Multiplex PCR Master Mix. The cycle was as follows: 98 °C for 3 min; followed by 10 cycles at 98 °C for 10 s, 55 °C for 5 s, and 68 °C for 5 s; and a final elongation step at 68 °C for 5 min.
Barcoded products of 16S rRNA ITS1/2 gene amplification from the same samples were pooled in a mass ratio of 10:3. Pooled products from the different samples were mixed equally and 1D ligation kits (SQK-LSK109; Oxford Nanopore) were used to construct sequencing libraries. Then, the library was sequenced using Oxford Nanopore MinION. TE buffer was run in each batch as a negative control throughout DNA extraction, target amplification, library construction and sequencing.
Bioinformatics analysis
Fast5 files generated by MinION were real-time base called and demultiplexed using Albacore v2.3.1. Low-quality reads (less than 7) were filtered. Porechop was used to trim the barcodes and adapters from the raw reads. Afterwards, the filtered sequencing reads were mapped to the reference databases downloaded from the 16S rDNA/ITS reference database maintained by NCBI FTP (ftp://ftp.ncbi.nlm.nih.gov/refseq/TargetedLoci) using Blast, and the taxonomy of each read was assigned according to the taxonomic information of the mapped subject sequence.
Statistical analysis
IBM SPSS Statistics Software Version 20 (SPSS, Inc, Chicago, Illinois, USA) was used for data analysis. Continuous variables like VA and diagnostic time were compared using a non-parametric, two-sided Wilcoxon rank sum test. The McNemar's test and the kappa statistic were used to compare the diagnostic positivity rates, the frequency of polymicrobial infection, and the frequency of treatment change between two methods. P value less than 0.05 was considered to be statistically significant.
Results
Basic information and clinical features of the patients
Demographic characteristics, disease course, clinical features, management details, and visual outcomes were demonstrated in Table 1. A total of 30 males and 13 females were involved in our study and the mean age was 54.86 ± 18.29 years. Most cases (23/43, 53.49%) occurred after penetrating ocular injuries from metal objects, sticks, pencils, or stones. 16 patients (16/43, 37.21%) developed severe ocular inflammation after ophthalmic surgeries (14 after cataract surgery, 1 after pterygium excision, and 1 after implantable collamer lens (ICL) implantation). 1 patient had chronic comorbid conditions of liver abscesses and was suspected of binocular endogenous endophthalmitis. The mean interval between the insult (surgery/ trauma/ infection) and manifestation of the injury was 7.26 ± 7.79 (range, 1–30 days).
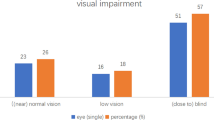
Presenting visual acuity (VA) results were as follows: no light perception (NLP; n = 2/44, 4.55%), light perception (LP; n = 9/44, 20.45%), hand motions (HM; n = 19/44, 43.18%), counting fingers (10/44, 22.73%), and undetermined VA (n = 4/44, 9.09%). These cases were characterized by severe anterior chamber inflammation and dense infiltration in the vitreous cavity. Hypopyon inflammatory/fibrinous exudation was observed in 33 of 44 eyes (75%). Visualization in the posterior segment view was poor in all 44 eyes due to severe vitritis (Fig. 1).
The visual outcome was defined as improvement, stabilization, and deterioration. Improved visual outcomes were found in 31 eyes (70.45%), and stabilized visual outcomes were found in 7 eyes (15.91%). 6 eyes (13.64%) had worse or deteriorated visual outcomes. 3 patients whose initial visions were LP, HM, and NLP respectively, underwent eye removal for severe inflammation involving the whole eye. Overall, the VA at the last follow-up was significantly improved compared with VA at the first visit (Z = −4.249, P < 0.001) (Table 2).
Clinical microbiology detected by NTS and culture
The results for NTS and culture are shown in Tables 3 and 4. In total, the positivity rate of NTS was 86.05% (37/43), and that of culture was 20.93% (9/43) (P < 0.001, kappa = 0.082). In all 9 culture-positive samples, the major pathogens were highly consistent with NTS results. Of the 37 NTS-positive samples, 22 showed polymicrobial infection. Whereas of the 9 culture-positive samples, only 1 showed polymicrobial infection. The frequency of polymicrobial results between NTS (22/43, 51.16%) and culture (1/43, 2.33%) was statistically significant (P < 0.001, kappa = 0.044).
Among 37 patients (38 eyes), a total of 45 species of bacteria and 11 species of fungi were identified by NTS, and no pathogens were detected in 6 eyes. These organisms were divided into pathogenic and non-pathogenic organisms. We have marked non-pathogenic organisms with an asterisk in Tables 3 and 4. The most frequently detected pathogens were Streptococcus spp., followed by Staphylococcus spp. and Enterobacter spp. Apart from well-recognized causative agents of endophthalmitis mentioned above, unusual and virulent pathogens were also revealed by NTS, including Sphingomonas paucimobilis, Mycobacterium abscessus, Stenotrophomonas maltophilia, Achromobacter spp., Aeromonas caviae, Morganella morganii, Acinetobacter junii, Colletotrichum spp., Moraxella osloensis, Bacillus cereus, Clostridium perfringens, Aeromonas veronii, and Citrobacter freundii.
A total of 20 microorganisms detected by NTS were identified as non-pathogenic. To our knowledge, some organisms have not been detected in any other clinical specimens: Cladosporium halotolerans, Paraburkholderia dipogonis, Yarrowia lipolytica, Sporidiobolus spp., Meyerozyma guilliermondii, Lactococcus spp., and Sac fungi. Whereas some were reported to colonize the oral cavity or the skin, and there were no reports of associated ocular infections caused by these organisms: Dialister spp., Micrococcus kristinae, Anaerococcus prevotii, Enterobacter cancerogenus, Corynebacterium tuberculostearicum, Atopobium parvulum, Corynebacterium jeikeium, Porphyromonas bennonis, Finegoldia magna, Corynebacterium confusum, Anaerococcus nagyae, Eubacterium tenue, and Lactococcus lactis.
All the patients obtained NTS results and determined treatment strategies within 1 or 2 days, with an average duration of 1.23 ± 0.43 days. The turnaround time for sequencing in the laboratory was around 8 h. The traditional culture required 3–4 days for bacterial detection and 5–7 days for fungal detection after collection of the specimens. The average turnaround time for culture was 3.58 ± 0.88 days. Thus, there was a significant difference between the two methods in terms of diagnostic time (Z = −5.805, P < 0.001). In this study, the mean duration of hospitalization in the Ophthalmic Center was 8.49 ± 2.45 days (range, from 1 to 16 days).
Management and changes in antibiotic strategy by the intervention of NTS
Due to the severity of the endophthalmitis, pars plana vitrectomy (PPV) was needed in 32 cases (74.42%). Each of these patients was first treated by a standard protocol with systemic and topical antibiotics, including intravenous and intravitreal ceftazidime (CEF)/vancomycin (VAN). After receiving NTS reports, 17 of 43 patients (39.53%) changed their antibiotic strategy. In contrast, only 5 of 43 patients (11.63%) were advised to change their medication after obtaining culture results. A significant difference existed in the medical guidance between the two methods (P < 0.001, kappa = 0.335).
Specifically, in patients #2, #31, #34, #35 and #38, virulent fungi including Aspergillus gracilis, Aspergillus penicillioides, Candida albicans, Colletotrichum spp., and Candida parapsilosis were detected. Then they received additional intravenous and intraocular voriconazole (VCZ)/gentamicin (Gen) immediately. For patient #8, in whom an emerging multidrug-resistant gram-negative bacilli Enterobacter ludwigii was detected, levofloxacin (LEV) was intravenously administered instead. In addition, high abundance of Aeromonas caviae was found in patient #29. This is a rare and destructive gram-negative bacterium that often lives in sewage and seawater. This patient worked for leech farming and lived in a humid environment, which confirmed the source of the pathogen. Therefore, we added CIP to her systemic antibiotics. The management was similar in patients #12, #24, #28, #33, #37, #39, #40, #41, and #43, and the details of changes in antibiotic strategies were shown in Table 5. Most of these patients responded favorably, but 3 patients underwent enucleation because of severe inflammation involving the whole eye.
NTS in endogenous endophthalmitis
Patient #4 with EE was presented with liver abscess, high fever and sepsis. Later he developed eye pains and vision loss in both eyes (Fig. 2). He was treated with intravenous MEM + VAN and binocular intraocular CEF + IPM, but no sign of symptom improvement was observed in the right eye. Then, Phaco + PPV + SOT + IVT was performed in his right eye to remove the inflammatory lesions. AH samples was collected in both eyes for NTS and microbiological culture. 24 h later, NTS revealed Stenotrophomonas maltophilia in the right eye, and Mycobacterium abscessus + Stenotrophomonas maltophilia in the left eye. Culture of AH samples revealed no pathogen. Subsequent cultures of liver abscesses also reported the presence of Klebsiella pneumoniae + Stenotrophomonas maltophilia + Mycobacterium abscessus, confirming the diagnosis of EE.
Chest X-ray and computed tomography images of patient #4. a Infected lesions in both lungs with bilateral pleural effusion, partially encapsulated. Inadequate expansion of lung tissue at the fluid surface and solid lung changes near the fluid surface. b Marked dilatation of the small intestine and acute intestinal obstruction due to inflammatory irritation. c Liver abscess (Indicated by a white arrow)
In view of the severe condition and the newly detected pathogens by NTS, we switched the systemic antibiotic regimen to “MEM + ETM + CIP + CLR + MH”. Meanwhile, puncture and drainage of liver abscess was performed. Surprisingly, both systemic and ocular symptoms were alleviated considerably. His final BCVA maintained 20/200 in the right eye and 20/20 in the left eye.
Discussion
The unique advantages of NTS
Since the ONT first released the MinION to early users in 2014, many proof-of-concept studies have demonstrated its applications in infectious disease diagnostics [5]. For instance, the surveillance of emerging infectious diseases outbreak [6], identification of pathogen drug resistance [, , 5, 7, 8], and disease-related microbial community characterization [9]. Other studies have also provided clinical examples of the validation of NTS for pathogen identification in various samples, including aqueous humor or vitreous fluid [10], blood [11], and nasopharyngeal swabs [12]. In the field of ophthalmology, NTS is ideal for the analysis of microorganisms in AH or VH since a very limited sample volume (0.2 mL) is sufficient for detection [10].
As a third-generation sequencing technology, NTS has two unique advantages over NGS. First, it exhibits higher species-level resolution through a long-read sequencing strategy, which enhances accuracy by avoiding mis-assembly of genomic repeat regions. Second, nanopore-based technology is considered real-time as the data are generated read by read, whereas NGS results are not available until the end of the sequencing run. NTS has the potential to detect microorganisms within minutes of starting sequencing [13] and provide reliable results within 6 h of sample receipt [14]. Therefore, it is particularly useful for early antibiotic administration through timely detection of pathogens.
Microbiology diagnostics by NTS
Similar to the study by Huang et al. [10], we were able to detect pathogens in intraocular fluid in a very short period of time with a high positivity rate. We also collated the culture and sequencing results of clinically suspected endophthalmitis from other researches in Table 6 (see Appendix A for full trans). Generally, there was a good correlation between NTS and standard culture results in double-positive cases. In this study, we also found that the main organisms identified by both methods were identical. However, the accuracy and sensitivity of the microbial profiles in culture results were poor. One potential explanation was frequent exposures to antibiotics before sample collection, which may have influenced bacterial cultivation. Also due to mutual inhibition mechanisms of bacteria, culture results often reported a single pathogen, indicating the risk of under-detection.
NTS technology has tremendous advantages in detecting multiple infections, especially in the case of mixed bacterial and fungal infections. In this article, polymicrobial results were reported in more than half of the patients by NTS. It allowed early identification of the uncultured and time-consuming microorganisms (e.g. anaerobes and fungi), regardless of prior use of broad-spectrum antibiotics [, 15, 16]. Therefore, NTS may function as a valuable supplementary to diagnostics when culture-based methods are flagged as negative.
NTS enables early targeted therapy
Optimal clinical decision-making depends on identifying clinically relevant organisms present in the sample. However, conventional culture methods are always too slow and often fail to identify unusual or fastidious organisms. The average waiting times for the results of bacterial and fungal cultures were 48 and 72 h respectively, which is not conducive to guiding targeted antimicrobial therapy, especially for ocular emergencies like endophthalmitis.
While in the case of NTS, even with atypical and low-abundance pathogens, the turnaround time from sample to result was no more than 24 h [, 17, 18]. Thus, NTS enables early targeted therapy by reducing detection time and clinical turnaround time. When atypical and virulent pathogens are detected and inadequate therapy is given, NTS may save vision and reduce the risk of blindness by altering antibiotic therapy without delay [, 15, 16]. When no pathogens are detected or the detected microorganisms are determined to be non-pathogenic, this approach may contribute to an early de-escalation of broad-spectrum therapy, delaying antimicrobial resistance (AMR) [19]. Noticeably, as with the EE case in our study, NTS may be instructive in both topical and systemic medication.
Due to the presence of the blood-retinal barrier (BRB), NTS-guided antibiotic therapy is an effective complementary to patient management, but not a substitute for surgical treatment when persistent vitritis occurs [20]. We noticed the persistent symptoms and poor VA outcomes despite the coverage of broad-spectrum antibiotics in some patients, and PPV was needed to remove purulent lesions. This may be explained by the fact that visual outcomes in endophthalmitis are related to several factors, including presenting visual acuity, the presenting interval, and the promptness of appropriate therapy. Thus, further studies are required to clarify the role of NTS in altering the course of the disease and improving long-term VA. However, it has been noted in an array of literature that although the role of surgical and medical treatment in endophthalmitis varies, the most important intervention remains immediate intravitreal antibiotic injection [21]. Meanwhile, considering that poor visual prognosis of endophthalmitis is strongly associated with the type of pathogens involved, the identification of causative pathogens may still have important implications in predicting visual prognosis in the early stage of the disease.
Data interpretations for NTS in a clinical setting
NTS is a hypothesis-free approach and it has the potential to detect any unknown DNA-based microorganism in a clinical sample. This not only offers the promise of improved detection of traditional organisms, but also the ability to identify organisms not previously associated with endophthalmitis.
Comprehensive description of the microbial constituents may provide additional benefits: For one thing, multiple pathogens including less-common ones are assessed simultaneously during the initial sequencing run, thereby avoiding many rounds of testing. For another, it allows in-depth investigations of the ocular microbial community. This is vital both in maintaining ocular homeostasis [22] and in the pathophysiology of the disease. Changes in the eye microbiome have been confirmed to be linked with disease states like dry eye, diabetic retinopathy, glaucoma, macular degeneration, and infectious keratitis [23]. In recent studies, nanopore sequencing was proposed to monitor changes in the gut microbiome over time [, 24, 25]. Likewise, NTS could be adopted for monitoring the ocular microbiome in real-time and even function as a prognostic tool for ocular infectious and inflammatory conditions when validated further [26].
However, as with any sequencing technique, it has its limitations in determining which organisms are merely colonizers or contaminants, rather than pathogenic organisms. In response to this issue, researchers have applied variable cutoff values (e.g. > 20 mapped read pairs per million read pairs (rM) [27], > 50 reads [28], > 10 reads per million (RPM) ratio metric [29] and > 500 reads [30]) to limit the over-interpretation of low abundance microorganisms. A recent study using single gene targeted nanopore sequencing provided evidence that the samples having < 20 reads generally had a low load of pathogen [31]. Similarly, in our study, the cut-off value for the positive diagnosis was 20 reads. Besides, we have segregated the pathogenic microorganisms from those that are known to be commensals (as shown in Tables 2 and 3). In summary, clinicians need to evaluate NTS results carefully and avoid antibiotic abuse.
Conclusion
In conclusion, by comparing culture and NTS results, and analyzing patients’ clinical-oriented aspects, we demonstrated the superiority of NTS in diagnosing and guiding early treatment of endophthalmitis. Based on previous studies, we expanded the sample size to further elucidate the role of the NTS technique in clinical settings. NTS has already shown great potential for clinical applications due to its features of long-read sequences and real-time analysis. It promises to be an exceptionally powerful supplementary to traditional culture methods.
References
Cho H, Shin YU, Siegel NH et al (2016) Endogenous Endophthalmitis in the American and Korean population: an 8-year retrospective study. Ocul Immunol Inflamm 26:496–503. https://doi.org/10.1080/09273948.2016.1195000
Leal SM, Rodino KG, Fowler WC, Gilligan PH (2021) Practical guidance for clinical microbiology laboratories: diagnosis of ocular infections. Clin Microbiol Rev 34:e00070-e119. https://doi.org/10.1128/CMR.00070-19
Zhang Y, Lu X, Tang LV et al (2023) Nanopore-Targeted Sequencing Improves the Diagnosis and Treatment of Patients with Serious Infections. mBio 28:e0305522. https://doi.org/10.1128/mbio.03055-22
Fu Y, Chen Q, Xiong M et al (2022) Clinical performance of nanopore targeted sequencing for diagnosing infectious diseases. Microbiol Spectr 10:e00270-e322. https://doi.org/10.1128/spectrum.00270-22
Charalampous T, Kay GL, Richardson H et al (2019) Nanopore metagenomics enables rapid clinical diagnosis of bacterial lower respiratory infection. Nat Biotechnol 37:783–792. https://doi.org/10.1038/s41587-019-0156-5
Hoenen T, Groseth A, Rosenke K et al (2016) Nanopore sequencing as a rapidly deployable ebola outbreak tool. Emerg Infect Dis 22:331–334. https://doi.org/10.3201/eid2202.151796
Ashton PM, Nair S, Dallman T et al (2015) MinION nanopore sequencing identifies the position and structure of a bacterial antibiotic resistance island. Nat Biotechnol 33:296–300. https://doi.org/10.1038/nbt.3103
Břinda K, Callendrello A, Ma KC et al (2020) Rapid inference of antibiotic resistance and susceptibility by genomic neighbour typing. Nat Microbiol 5:455–464. https://doi.org/10.1038/s41564-019-0656-6
Hardwick SA, Chen WY, Wong T et al (2018) Synthetic microbe communities provide internal reference standards for metagenome sequencing and analysis. Nat Commun 9:3096. https://doi.org/10.1038/s41467-018-05555-0
Huang Q, Fu A, Wang Y et al (2021) Microbiological diagnosis of endophthalmitis using nanopore targeted sequencing. Clin Exp Ophthalmol 49:1060–1068. https://doi.org/10.1111/ceo.13992
Li JY, Shen GG, Liu TG et al (2021) Nanopore-targeted sequencing for simultaneous diagnosis of suspected sepsis and early targeted therapy. Ann Transl Med 9:1749. https://doi.org/10.21037/atm-21-2923
Wang M, Fu A, Hu B et al (2020) Nanopore targeted sequencing for the accurate and comprehensive detection of SARS-CoV-2 and other respiratory viruses. Small 16:e2002169. https://doi.org/10.1002/smll.202002169
Stoddart D, Heron AJ, Mikhailova E et al (2009) Single-nucleotide discrimination in immobilized DNA oligonucleotides with a biological nanopore. Proc Natl Acad Sci USA 106:7702–7707. https://doi.org/10.1073/pnas.0901054106
Greninger AL, Naccache SN, Federman S et al (2015) Rapid metagenomic identification of viral pathogens in clinical samples by real-time nanopore sequencing analysis. Genome Med 7:99. https://doi.org/10.1186/s13073-015-0220-9
Zhang R, Hu L, Xu C et al (2021) Bordetella avium-associated endophthalmitis: case report. BMC Infect Dis 21:833. https://doi.org/10.1186/s12879-021-06546-1
Liu C, Zhang L, Liu L et al (2022) Case report: first case of endophthalmitis caused by an emerging pathogen: nocardia huaxiensis. Front Publ Health 10:933851. https://doi.org/10.3389/fpubh.2022.933851
Kai S, Matsuo Y, Nakagawa S et al (2019) Rapid bacterial identification by direct PCR amplification of 16S rRNA genes using the MinION nanopore sequencer. FEBS Open Bio 9:548–557. https://doi.org/10.1002/2211-5463.12590
Jun KI, Oh BL, Kim N et al (2021) Microbial diagnosis of endophthalmitis using nanopore amplicon sequencing. Int J Med Microbiol 311:151505. https://doi.org/10.1016/j.ijmm.2021.151505
Trotter AJ, Aydin A, Strinden MJ, O’Grady J (2019) Recent and emerging technologies for the rapid diagnosis of infection and antimicrobial resistance. Curr Opin Microbiol 51:39–45. https://doi.org/10.1016/j.mib.2019.03.001
Sachdeva MM, Moshiri A, Leder HA, Scott AW (2016) Endophthalmitis following intravitreal injection of anti-VEGF agents: long-term outcomes and the identification of unusual micro-organisms. J Ophthalmic Inflamm Infect 6:2. https://doi.org/10.1186/s12348-015-0069-5
Peck TJ, Patel SN, Ho AC (2021) Endophthalmitis after cataract surgery: an update on recent advances. Curr Opin Ophthalmol 32:62–68. https://doi.org/10.1097/ICU.0000000000000727
Ozkan J, Willcox MD (2019) The ocular microbiome: molecular characterisation of a unique and low microbial environment. Curr Eye Res 44:685–694. https://doi.org/10.1080/02713683.2019.1570526
Cavuoto KM, Banerjee S, Galor A (2019) Relationship between the microbiome and ocular health. Ocul Surf 17:384–392. https://doi.org/10.1016/j.jtos.2019.05.006
Sepsis Lung Microbiome Study Group (2020) Could lung bacterial dysbiosis predict ICU mortality in patients with extra-pulmonary sepsis? A proof-of-concept study. Intensive Care Med 46:2118–2120. https://doi.org/10.1007/s00134-020-06190-4
Leggett RM, Alcon-Giner C, Heavens D et al (2020) Rapid MinION profiling of preterm microbiota and antimicrobial-resistant pathogens. Nat Microbiol 5:430–442. https://doi.org/10.1038/s41564-019-0626-z
Ciuffreda L, Rodríguez-Pérez H, Flores C (2021) Nanopore sequencing and its application to the study of microbial communities. Comput Struct Biotechnol J 19:1497–1511. https://doi.org/10.1016/j.csbj.2021.02.020
Doan T, Wilson MR, Crawford ED et al (2016) Illuminating uveitis: metagenomic deep sequencing identifies common and rare pathogens. Genome Med 8:90. https://doi.org/10.1186/s13073-016-0344-6
Wang J, Han Y, Feng J (2019) Metagenomic next-generation sequencing for mixed pulmonary infection diagnosis. BMC Pulm Med 19:252. https://doi.org/10.1186/s12890-019-1022-4
Miller S, Naccache SN, Samayoa E et al (2019) Laboratory validation of a clinical metagenomic sequencing assay for pathogen detection in cerebrospinal fluid. Genome Res 29:831–842. https://doi.org/10.1101/gr.238170.118
Heikema AP, Horst-Kreft D, Boers SA et al (2020) Comparison of illumina versus nanopore 16S rRNA gene sequencing of the human nasal microbiota. Genes (Basel) 11:1105. https://doi.org/10.3390/genes11091105
Zhou L, Lopez Rodas A, Llangarí LM et al (2022) Single gene targeted nanopore sequencing enables simultaneous identification and antimicrobial resistance detection of sexually transmitted infections. PLoS One 17:e0262242. https://doi.org/10.1371/journal.pone.0262242
Zhu J, Xia H, Tang R et al (2022) Metagenomic next-generation sequencing detects pathogens in endophthalmitis patients. Retina 42:992–1000. https://doi.org/10.1097/IAE.0000000000003406
Low L, Nakamichi K, Akileswaran L et al (2022) Deep metagenomic sequencing for endophthalmitis pathogen detection using a nanopore platform. Am J Ophthalmol 242:243–251. https://doi.org/10.1016/j.ajo.2022.05.022
Mishra D, Satpathy G, Chawla R et al (2021) Targeted metagenomics using next generation sequencing in laboratory diagnosis of culture negative endophthalmitis. Heliyon 7:e06780. https://doi.org/10.1016/j.heliyon.2021.e06780
Selva Pandiyan A, Siva Ganesa Karthikeyan R, Rameshkumar G et al (2020) Identification of bacterial and fungal pathogens by rDNA gene barcoding in vitreous fluids of endophthalmitis patients. Semin Ophthalmol 35:358–364. https://doi.org/10.1080/08820538.2020.1864416
Kosacki J, Boisset S, Maurin M et al (2020) Specific PCR and quantitative real-time PCR in ocular samples from acute and delayed-onset postoperative endophthalmitis. Am J Ophthalmol 212:34–42. https://doi.org/10.1016/j.ajo.2019.11.026
Deshmukh D, Joseph J, Chakrabarti M et al (2019) New insights into culture negative endophthalmitis by unbiased next generation sequencing. Sci Rep 9:844. https://doi.org/10.1038/s41598-018-37502-w
Gandhi J, Jayasudha R, Naik P et al (2019) Targeted high-throughput sequencing identifies predominantly fungal pathogens in patients with clinically infectious. C Negat Endophthalmitis South India Microorg 7:411. https://doi.org/10.3390/microorganisms7100411
Mishra D, Satpathy G, Chawla R et al (2019) Utility of broad-range 16S rRNA PCR assay versus conventional methods for laboratory diagnosis of bacterial endophthalmitis in a tertiary care hospital. Br J Ophthalmol 103:152–156. https://doi.org/10.1136/bjophthalmol-2018-312877
Pongsachareonnont P, Honglertnapakul W, Chatsuwan T (2017) Comparison of methods for identifying causative bacterial microorganisms in presumed acute endophthalmitis: conventional culture, blood culture, and PCR. BMC Infect Dis 17:165. https://doi.org/10.1186/s12879-017-2264-5
Lee AY, Akileswaran L, Tibbetts MD et al (2015) Identification of torque teno virus in culture-negative endophthalmitis by representational deep DNA sequencing. Ophthalmology 122:524–30. https://doi.org/10.1016/j.ophtha.2014.09.001
Bharathi MJ, Murugan N, Rameshkumar G et al (2013) Comparative evaluation of uniplex, nested, semi-nested, multiplex and nested multiplex PCR methods in the identification of microbial etiology of clinically suspected infectious endophthalmitis. Curr Eye Res 38:550–62. https://doi.org/10.3109/02713683.2013.772205
Chiquet C, Cornut PL, Benito Y et al (2008) Eubacterial PCR for bacterial detection and identification in 100 acute postcataract surgery endophthalmitis. Invest Ophthalmol Vis Sci 49:1971–8. https://doi.org/10.1167/IOVS.07-1377
Chiquet C, Lina G, Benito Y et al (2007) Polymerase chain reaction identification in aqueous humor of patients with postoperative endophthalmitis. J Cataract Refract Surg 33:635–41. https://doi.org/10.1016/j.jcrs.2006.12.017
Funding
This study was supported by the Natural Science Foundation of Hubei Province (grant no. 2019CFB489).
Author information
Authors and Affiliations
Contributions
All authors contributed to design of the study, data collection, analysis, interpretation, drafting, and revision of the paper. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare.
Ethics approval
This study was carried out following the institutional guidelines and ethical standards of the 1964 Declaration of Helsinki and was approved by the Institutional Review Board of Renmin Hospital of Wuhan University (WDRY2019-K056).
Consent to participate
Informed consent was obtained from all individual participants included in the study.
Consent for publications
The authors affirm that human research participants provided informed consent for publication of the images in Figs. 1a, b, c, d, 2a, b, c. Informed consent was obtained from Patient 4 to publish potentially identifying information, such as details and photographs of the case.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix A
Appendix A
See Table 6.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, X., Li, Z., Wang, M. et al. The diagnostic utility of nanopore targeted sequencing in suspected endophthalmitis. Int Ophthalmol 43, 2653–2668 (2023). https://doi.org/10.1007/s10792-023-02665-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10792-023-02665-7