Abstract
This paper presents optimization methods for the K-means segmentation algorithm for a sequence of thermal images. Images of the sample response in the frequency domain to the thermal stimulation with a known spectrum were subjected to cluster segmentation, grouping pixels with similar frequency characteristics. Compared were all pixel characteristics in the function of the frame number and grouped using the minimal sum of deviations of the pixels from their segment mean for all the frames of the processed image sequence. A new initialization method for the K-means algorithm, using density information, was used. A K-means algorithm with a kd-tree structure C# implementation was tested for speed and accuracy. This algorithm divides the set of pixels to the subspaces in the hierarchy of a binary tree. This allows skipping the calculation of distances of pixels to some centroids and pruning a set of centroid clusters through the hierarchy tree. Results of the segmentation were compared with the K-means and FCM algorithm MATLAB implementations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
As a result of thermal excitation, a solid gives us a temperature response that we can study in both time and frequency domains. The sequence of thermal images includes in successive frames various information, depending on the internal structure of the sample. Analysis of thermal image sequences provides information about the quality of a sample’s inner layers, its defects, and changes of thermal parameters. It can be used for detection of very small substance concentrations, study of photochemical reactions (e.g., photosynthesis), or obtaining depth profiles of samples in thermal microscopy.
Changes in the thermal sequence frames are continuous, and we can talk about the characteristics of each pixel in the sequence. To reduce processing, one can use cluster segmentation, which has a well-defined error and allows for an accurate quality comparison of various algorithms used for this purpose. This feature is quite rare among segmentation methods.
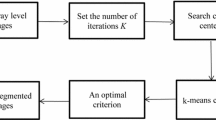
This paper presents a method of optimizing a K-means algorithm for segmentation of thermal image sequences using a kd-tree structure, which divides a set of the pixels to subspaces in the hierarchy of a binary tree. This allows skipping of the pixels distance calculation to some centroids and pruning clusters from the set of clusters during a search of the hierarchy tree. A new method of K-means algorithm initialization, using information about the pixels density in a sequence, was applied. Results of the speed and accuracy of the segmentation algorithm were compared with the implementation of the K-means algorithm and FCM algorithm implementations in MATLAB.
2 Principles of Algorithms
2.1 Description of Cluster Segmentation Algorithms
The fastest and most popular cluster segmentation algorithm is K-means (Forgy [1], MacQueen [2], Lloyd [3], Wu et al. [4]). Image clustering using K-means is a pixel segmentation algorithm, depending only on pixel values in consecutive frames and not taking into account locations and values of the neighbor pixels. The result of the segmentation consists generally of k incoherent objects, composed of pixels that are not necessarily located in their neighborhood, but of similar values. The procedure for the segmentation of an image sequence determines the sets of similar pixel value characteristics in a function of the frame number. From now on, for simplicity, we will in this article understand the term pixel as a vector of pixel values in successive frames of the sequence. The input parameter of the procedure is a set of k vectors of initial clusters. To eliminate the possibility of assigning two similar pixels to two different clusters, after assignment of pixels to the nearest segments, cluster centroids are recalculated to the mean value from all the pixels belonging to the cluster, and the assignment step is repeated (Lloyd algorithm). The iteration is interrupted when the pixels cease to change their assignment to clusters. The K-means algorithm is always convergent if the measure of the distance vectors is the Euclidean metric \(L_{2}\), but the speed of convergence and accuracy of local solutions is determined mainly by the choice of the initial vector clusters (Pena et al. [5]). The standard solution is to choose centroids at random with a uniform probability distribution from all the values in the set (Forgy approach [1]). A KKZ method (Katsavounidis et al. [6]) is also a fast seeding algorithm in which pixels farthest from existing centroids are chosen for the new centroids. This prevents dividing a cluster into several smaller ones in case of choosing centroids, which are close to each other. Unfortunately, this method is very sensitive to outliers in a sequence. Arthur and Vassilvitskii in [7] present a K-means++ algorithm, in which the initial centroids are selected from a set of input pixels, assuming a pixel probability proportional to the distance from the found centroids. The method thus combines the use of statistics and analysis of the mutual distance of clusters. The most common initialization algorithms for K-means, however, do not take into account the density distribution of pixels. Use of such information and the kd-tree structure is presented by Redmond and Heneghan [8]. Determination of the density distribution requires calculation of the mutual distances between pixels, which needs O(\(n^{2})\) count of operations. For a pixel k, its density can be determined, for example, by using the sum of the inverses of the pixel distances to all other pixels. The distance to near pixels has a much greater influence on this function value than to far ones (similar to the inverse distance weighting function). Factor \(\varepsilon \) prevents the function from escaping to infinity for cases, when \(x_i =x_k \).
Factor \(\varepsilon \) corresponds to a resolution of recorded distances; hence, it should satisfy \(\varepsilon \ll \left( {x_i -x_k } \right) ^{2}\) for all \(i\), except for the case: \(x_i =x_k \).
Using a Taylor series expansion of the function \(\left( {t+1} \right) ^{-1}=1-t+t^{2}-t^{3}+\cdots \) and expanding every term in the sum (Eq. 1) for \(t=\frac{\left( {x_i -x_k } \right) ^{2}}{\varepsilon }\), we obtain
The series converges only when \(\varepsilon >\left( {x_i -x_k } \right) ^{2}\) for each pair of \(i,k\).
Let us denote the vector of mean values of all pixels as \(\overline{x}\) and the variance of pixel’s values as
The density (Eq. 1) could be estimated, taking distances to be equal to the fixed mean value \(d_{k}\):
Expanding both sides of the equation into a Taylor series,
and comparing second terms of both sides, \(d_{k}\) can be evaluated as
Since \(\varepsilon \) should be as small as possible, we want to choose for its value the convergence limit for the series \(\varepsilon \approx d_k \). Additionally, \(\varepsilon \) has to be the same for all pixels, so we take the mean value of all \(d_{k}\):
And finally we can estimate the pixel’s \(k\) density as
The local pixel density approximation using Eq. 8 requires O(\(n)\) operations.
For the segment centroids, the pixels with a greater local density, but also more distant from the already found centers should be selected. In our experiments, as the first center, the pixel with the highest density is selected. Due to the less accurate estimation of the local density, the other centers of segments were chosen from pixels with a maximum product of the square root of the density and its distance from the already found centers. We denote such an initialization method as high density (HD) for the density described by Eq. 1 and its approximation using Eq. 8.
The segmentation algorithm Fuzzy C-Means (FCM, Bezdek [9]) assumes that the pixels may be partially assigned to a segment as defined by the membership function \(\mu \) (soft membership). Equation 9 presents a modification of cluster centroids to the center of gravity of the segments for each frame, which takes place after iteration:
Equation 10 defines the membership function for each pair of the ith pixel and jth segment:
where r is the fuzziness of the member function, \(r\ge \) 1 and S is the segment count.
The K-means algorithm is thus a special case of the FCM method, for which the membership function is binary, taking one for the pixel and assigned segment pair and zero for the pixel with other segments (hard membership). The FCM algorithm can find, in general, a better solution, resulting, however, with increased computational effort and hence, a longer processing time.
The standard implementation of the K-means algorithm calculates the distances between pixels and all the cluster centers. As long as the cluster centers change, distance calculations for pixels have to be evaluated, and pixels are assigned to the nearest segments. However, organizing the collection of pixels in a kd-tree structure, one can construct a K-means algorithm which assigns groups of pixels to the segment simultaneously, and not one segment to one pixel at a time. There is often no need for calculating distances from each pixel to the centers of each segment. Groups of pixels can be assigned to a segment, or rejected as located too far away. The input data are two-dimensional (pixel values as a function of the frame number). The kd-tree could be created by comparing the data alternately in these dimensions. More effective, however, is to sort characteristics, for example, on their mean values (reducing the data to a single dimension). A more accurate similarity indicator is the square length of the characteristic vector. Since the square length of the vector is always non-negative, in order to distinguish the characteristics of a negative mean value, it is assumed that the similarity index is the square length of the vector with the sign from the mean value. Such a classification groups similar characteristics in the nodes of the kd-tree, fewer comparisons during processing are needed, and the computation time is shorter. Equation 11 shows how to determine the characteristic square length Len of the pixel vector v (with a dot product), by which pixels are sorted. Vector ones has coordinates all equal to 1.
On a sorted pixels basis, a binary kd-tree is created. The leaves of the tree correspond to the pixels.
The new nodes are created from two nodes of the lower level, starting from the lowest level of the leaves. In each node the maximum distance of its children from the center is stored—the node deviation. During segmentation, the kd-tree is searched recursively using a filtering procedure. The parameter for the procedure is a list of clusters, in which segments located too far away from the center of the node are pruned (Fig. 1). The triangle inequality for distances is used. The distance between a segment and any node’s pixel is less than a sum of distance \(\varDelta \) from a segment to a node and the node’s deviation d, but greater than the difference between them. The segment is close enough to the node’s pixels if its minimum distance to the pixels will be smaller than the maximum distance between the pixels of the current node and its nearest segment. In Eq. 12 \({\varDelta }_{N}\) is the distance from the node to its nearest segment (Fig. 2);
Pseudocode of filtering procedure is presented below:

The node-to-segment assignment involves updating the sum of segment centroids and segment centroids count by summing with the node’s respective properties. This enables a later calculation of the segment centroid without aggregating error.
When using the Euclidean norm \(L_{2}\), taking a triangle inequality into account requires the calculation of square roots of vector squared lengths (inequality for the squared lengths may not be satisfied). Such problems are not posed by city metric \(L_{1}\) (Manhattan, taxi), but K-means using \(L_{1}\) and the algorithm is not always convergent. The proper method would determine centers of segments based on the median pixels rather than the mean (K-median algorithm). We do not use correlation in our calculation either, because correlation indicates the degree of similarity between the characteristics, but does not allow determination of the characteristic most similar to all others characteristics of pixels in the segment.
In general, during segmentation, some segments can eventually lose all their pixels. If we choose for the initial centers pixel values from the sequence, it will mean that some of the selected pixels must have identical characteristics. The algorithms implemented in MATLAB use two strategies—ignore (drop) and selecting for the center of the empty segment a pixel which is farthest from its center (singleton). In our implementation of the K-means, if the initial centers of segments are selected from pixels in sequence, none of the segments ever lose all their pixels, because at least one pixel is at zero distance from a segment’s center, and a pixel check of the segment-to-pixel distance starts from the currently assigned segment. In this way we eliminate the need for special processing of empty segments. This technique is also used in an algorithm with the kd-tree.
2.2 Used Algorithms and Techniques
Clustering algorithms in MATLAB were performing a batch update phase only, without trying to improve the result with a time-consuming on-line phase. In the batch update mode, if the segment has not lost pixels, or gained new ones, calculations for its distance to the pixels were not repeated. In this way, successive iterations are performed faster and the segmentation process is significantly shortened. In our implementation, in addition to memorizing segments, whose centers have not been moved, the distances of pixels from the segments, which are known to be changed in the next iteration, are calculated only until reaching the excess of the minimum currently known distance between the pixel and the segment. Additionally, an algorithm was developed in which adding or removing pixels from a segment is reflected immediately, not after the iteration (dynamic update).
In the experiments we used algorithms K-means and FCM in MATLAB, object-oriented algorithms developed in C# with the update of the segments at the end of an iteration or with a dynamic update and the K-means algorithm using the kd-tree structure.
2.3 Segmentation Error
For segmentation of an image sequence, the absolute error for S objects is defined as the sum of deviations of all P pixels from the nearest centers of segments for all K frames of the sequence:
The absolute error can also be presented as the sum of the absolute errors in individual segments and illustrated as the characteristic of the absolute segment errors in a function of the segment number:
3 Results
We show the study of three sequences of thermal images with a resolution of \(128\times 128\) pixels and the number of frames from 1 to 512 (Fig. 3). The K-means algorithm has a very strong dependence on the choice of the initial centers of the segments. Therefore, in the first experiment, we compared the HD initialization method with Forgy’s approach for 20 iterations, KKZ, and the HD approximation method. Comparison of methods for initializing the K-means algorithm is shown in Table 1. Selection of the initial centers determines the error of the first iteration. That error for this experiment was also given, as a small final error is related to finding a good initial estimate. A small initial error cannot guarantee, and is not sufficient to achieve a good end result, but as experiments reveal, it is a prerequisite. Results of different initialization methods are compared in Fig. 4 with the results of 20 consecutive random initializations. The HD initialization got the best results. An approximation for the HD method also obtained better results than for the KKZ and FA initializations. Due to the high computational complexity of the HD method, in further experiments, we used the HD approximation, calculating the density of pixels in accordance with Eq. 8. Prior to segmentation using the kd-tree, the pixels were sorted according to their characteristic length.
Table 2 present results of the segmentation of a Seq2 sequence for a different number of frames using all investigated K-means algorithms. Figure 5 illustrates this as calculation time function plots. The FCM algorithm is computationally much more complex than KM; it is difficult to converge and requires choosing an appropriate fuzziness factor r for every change in the number of processed frames of the sequence. In our experiment, we adopted the constant r = 1.5 and the number of iterations is equal to 200. It is worthwhile to mention that the C# code does not produce a native code, and therefore is slower than an application written in C++, but it is relatively easy in a C# environment to implement parallel processing using a parallel. For loop, the computation time was therefore shorter than the MATLAB implementation by a factor proportional to the number of processor cores—in our case, twice. Dynamic update segmentation converges faster than other algorithms. The computation time is longer only because it does not use parallel processing of pixels. Changing the order of pixel processing would affect, in this case, the final result of segmentation.
Figure 6 presents a result image of Seq1 sequence segmentation in which pixel values are equal to the mean value of the associated segment’s center vector. Using a jet palette for pseudocolors, the correctness of segmentation could be easily verified. Four segments from this image are chosen, and all pixel characteristics for a given segment are presented in images in Fig. 7.
All algorithms implemented in C# are object-oriented, enabling easy analysis and code modification. Implementation of other segmentation algorithms, such as hierarchical segmentation, is planned in the future.
4 Conclusion
This paper presents a new method for initializing a K-means algorithm, which takes into account the density of pixels in a sequence. A quick estimate method for calculating the density of pixels with a complexity of O(\(n)\) has also been presented. Using this additional information, the HD method obtains smaller segmentation errors than KKZ and FA initializations.
K-means segmentation using the kd-tree has proved to be very fast with segmentation of single images, but segmentation of the sequences takes much longer and successive iterations are not performed as fast as in standard implementations of a K-means algorithm. With sorting of one-dimensional data, a K-means algorithm using the kd-tree can be used to segment individual images in real time. Due to the recursive nature of the algorithm, there are considerable difficulties in accelerating consecutive iterations of the algorithm and parallelization of computations.
Sequences of thermal images are contained in each pixel of the frame, temperature information associated with the same location on the sample surface but also with the depth information associated with the frame number. This example of specific thermal image sequences enables very high information compression using cluster segmentation. As a result, pixels with similar thermal properties are combined into groups—clusters. Such a reduction in quantity of information can allow real-time implementation of thermal tomography.
References
E. Forgy, Biometrics 21, 768 (1965)
J. MacQueen, in Proceedings of the 5th Berkeley Symposium on Mathematical Statistics and Probability, vol. 1 (University of California Press, 1967), p. 281
S.P. Lloyd, IEEE Trans. Inf. Theory. 28, 129 (1982)
X. Wu, V. Kumar, J.R. Quinlan, J. Ghosh, Q. Yang, H. Motoda, G.J. McLachlan, A.F.M. Ng, B. Liu, P.S. Yu, Z. Zhou, M. Steinbach, D.J. Hand, D. Steinberg, Knowl. Inf. Syst. 14, 1 (2008)
J.M. Pena, J.A. Lozano, P. Larranaga, Pattern Recognit. Lett. 20, 1027 (1999)
I. Katsavounidis, C. Kuo, Z. Zhang, IEEE Signal Process. Lett. 1, 144 (1994)
D. Arthur, S. Vassilvitskii, in Proceedings of the 18th annual ACM-SIAM Symposium on Discrete Algorithms (2007), p. 1027
S.J. Redmond, C. Heneghan, Pattern Recognit. Lett. 28, 965 (2007)
J.C. Bezdek, Pattern Recognition with Fuzzy Objective Function Algorithms (Plenum Press, New York, 1981)
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Świta, R., Suszyński, Z. Cluster Segmentation of Thermal Image Sequences Using kd-Tree Structure. Int J Thermophys 35, 2374–2387 (2014). https://doi.org/10.1007/s10765-014-1688-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10765-014-1688-z