Abstract
The subterranean aquatic snails may serve as a model of endemism and isolation vs. migration in subterranean habitats. The aim of the present paper is to verify the hypothesis that subterranean aquatic snails can migrate through diverse subterranean habitats, applying four molecular markers as well as a RAPD technique and shell morphometry. They were used to estimate the differences and gene flow between populations of the hydrobiid subterranean aquatic species Montenegrospeum bogici, collected in the Dinaric karst region. Three molecularly distinct taxonomic units were distinguished. The mOTU B was found at single locality, mOTU C at two, but the mOTU A at ten localities, scattered along 236 km distance, at two of them in sympatry with either mOTU B or C. Within mOTU A, the estimated levels of the gene flow were high. The pairwise measures of genetic differentiation were statistically significantly associated with geographic distances between the populations. In general, neither the infinite-island model of interpopulation differentiation, expected for isolated populations, nor the stepping-stone one, but rather the isolation-by-distance model explained the observed pattern. Our results suggest that interstitial habitats provide ways of migration for the stygobiont M. bogici, as has been already suggested for other subterranean gastropods.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Of the approximately 20,000 worldwide described species of subterranean animals (Culver & Pipan, 2009), stygobionts and troglobionts, there are over 350 described species of stygobiont (obligate subterranean-dwelling aquatic) gastropods, with 97% of them, and over 50 genera representing the superfamily Truncatelloidea (“Hydrobiidae s. lato” in Bernasconi & Riedel, 1994; Culver, 2012). They are worldwide distributed in all kind of brackish and freshwater habitats, and grouping small or minute (shell height 1–3 mm), dioecious gill-breathing snails, whose phylogenetic relationships are still far from being clear, and whose systematics remains poorly understood. Sket et al. (2004) reported 169 stygobiont gastropod species inhabiting the Balkan Peninsula; 148 of them, representing 41 genera and 6 families, were found in the Western Balkans, 8 (4 truncatelloid genera) in Greece, and 13 (11 truncatelloid genera) in Bulgaria. Among the subterranean gastropods, aquatic species are much more numerous than the terrestrial ones—for example, in the USA stygobionts’ species diversity is nearly six times higher than that of troglobionts (Hobbs, 2012; Gladstone et al., 2021). However, despite the obvious existence of many still undescribed species in these not easily accessible and explorable habitats, the number of those already known is overestimated (e.g. Falniowski & Beran, 2015; Osikowski et al., 2018). As pointed out by Falniowski (2018), using intraspecies variable shells as the only known structures, coupled with the widespread belief among many taxonomists in geographic isolation and unavoidable, immediate speciation of the cave animals has resulted in descriptions of new species (nearly) in each cave or other subterranean habitat (e.g. Reischütz et al., 2008; Georgiev, 2013; Glöer et al., 2015; Grego et al., 2017). The incredibly long list (more than 60 species and many subspecies) of the nominal species of Bythiospeum Bourguignat, 1882 presented by Glöer (2002), may serve as a good example. Even after including detailed, thorough morphological (e.g. shell morphology) and molecular (e.g. allozymes, mitochondrial data) information, the correct species identification in the Truncatelloidea still pose quite a challenge even for experienced taxonomic specialists (Falniowski, unpublished data). Usually one can find varying quantities of empty, often more or less damaged shells, found at the surface, at groundwater-fed springs. Among such shells only a few living individuals were ever found, washed out into springs at times of high flow, especially during the spring season (Haase, 1995; Richling et al., 2016). At a few localities, more living snails can sometimes also be found in springs and streams at the surface. There are also subterranean snails whose shells can only be found deep inside caves and are often incomplete, to say nothing of the extreme scarcity of living specimens. Collection of stygobiont Truncatelloidea is, therefore, difficult, especially in adequate numbers for any study of population genetic structure—subterranean populations are usually not dense, rich in individuals (e.g. Mammola et al., 2021). In fact, nothing but shell morphology is known for the vast majority of the subterranean gastropods. Even the soft parts, if accessible, are not usually informative enough to resolve taxonomic questions, since the animals are very tiny compared to other gastropods, and miniaturisation, coupled with convergent adaptations to freshwater habitats (osmoregulation, internal fertilisation, eggs in capsules, etc.) has resulted in simplification and unification of their anatomy (e.g. Fretter & Graham, 1994; Culver, 2012; Falniowski, 2018). Such incompleteness of the data, coupled with assumptions that they must be endemic, has resulted in strongly biased estimates of biodiversity, to say nothing about systematics, evolution, and ecology of these snails. Molecular data—DNA sequences—are helpful, but there are still only a few studies applying them (e. g. Grego et al., 2019; Hofman et al., 2019). Another limitation concerns the low number of applied molecular markers—in fact often COI only—which is a result of common problems with PCR amplification or lack of polymorphism. Many of the known stygobiont species primarily inhabit the interstitial habitats formed by the alluvial gravel (frequently not dependent on karst); thus, caves and spring are for them a secondary or occasional habitat, so their distribution ranges may be more widespread than originally supposed (Richling et al., 2016).
A relatively high species diversity of the subterranean snails could be expected, as the group is well preadapted to such an environment, as being nocturnal (or at least avoiding direct sunlight), opportunistic, ecologically (especially feeding) generalist animals, with well-developed mechano-sensory and/or chemo-sensory traits (Trajano & Cobolli, 2012). They are also well adapted to food-limited habitats, since their size-related energy requirements are low, being four times lower than in fishes and salamanders, and more than twice lower than in arthropod detritivores (Poulson, 2012). Thus, among the subterranean aquatic invertebrates, Crustacea dominate, but gastropods are the second group in species diversity (Culver & Sket, 2000). Animals adapted to subterranean, especially cave environments are often thought to be highly geographically isolated because of their limited dispersal ability, resulting from limited physiological tolerances and, especially in the case of snails, physical limitations of their locomotion (Purchon, 1977; Trueman, 1983). Nevertheless, the stygobiont gastropods (whose interstitial representatives hardly differ from the ones inhabiting caves) are known nearly only from taxonomic descriptions, mostly based on their shell morphology alone.
For example, Sbordoni et al. (2012) in their review of the structural studies of subterranean (meta) populations do not present any examples concerning the Gastropoda, as is also the case with Mammola et al. (2021). Simply there are no data about the Gastropoda from shallow hypogean habitats of many kinds, in similarity with hyporheic and deeper interstitial habitats along rivers and streams (Culver & Pipan, 2014). Apart from the descriptions of new species and faunistic lists, when considering the information on the stygobiont gastropod (meta)population genetic structure, robustly inferred levels of endemism, gene flow, etc. (perhaps because of the limited available material), the data are scarce. Thus, neither species boundaries nor phylogenetic relationships with the snails inhabiting surface environments are well understood (Falniowski, 2018). There are a few studies concerning the stygobiont gastropods, nearly exclusively of the Truncatelloidea. Falniowski et al. (1998, 1999) studied gene flow and metapopulation genetic structure (applying allozyme data) in Bythinella Moquin-Tandon, 1856 inhabiting springs. Bichain et al. (2007) also studied Bythinella in two caves (Padirac and Folatière) in France. Cytochrome oxidase (COI) sequences indicated several subterranean lineages, still belonging to the epigean species, thus, confirming that colonizations of hypogean habitats by epigean individuals are not rare events in the genus, as already suggested earlier (Boeters, 1979; Giusti & Pezzoli, 1982; Bole & Velkovrh, 1986; Bernasconi, 2000). Bichain et al. (2007) also found in sympatry or parapatry with those epigean lineages also distinct, exclusively hypogean phylogenetic lineages, thus, indicating multiple invasions of the caves and/or radiation already in caves. It has to be noted that Bythinella is, in general, not a stygobiont, but rather a stygophile, which means inhabiting both subterranean and epigean waters. In the western Balkans, Bythinella rather avoids the cave environment (JG personal observation).
COI sequences were used to estimate the time of divergence between Heleobia dobrogica (Grossu et Negrea, 1989) from the Movile Cave in Dobrogea, Romania and the epigean H. dalmatica (Radoman, 1974). The time 2.172 ± 0.171 Mya coincided with the period marking the beginning of the fall in temperature and precipitation that predated the Pleistocene, when H. dobrogica found a safe shelter within a warm cave (Falniowski et al., 2008). Falniowski & Sarbu (2015) described new species of Iglica A. J. Wagner, 1928 and Daphniola Radoman, 1973 from the Melissotripa Cave in Greece, inferring their molecular relationships (COI, 18S) with epigean species. Osikowski et al. (2015) studied the genetic and morphological differences between Bulgarian surface- and cave-dwelling populations of the genus Bythinella; while Rysiewska et al. (2016) have studied 16 populations of the Bulgarian cave/spring genus Pontobelgrandiella Radoman, 1978. Osikowski et al. (2017) described a new and entirely stygobiont clade from Bulgaria. Based on shell morphology and soft parts morphology and anatomy only, Hershler & Holsinger (1990) described the zoogeography of the stygobiont North American Truncatelloidea, demonstrating that in some regions of the USA and Mexico, some stygobiont hydrobioids originated from their ancestors invading cave habitats directly from the sea during the late Cretaceous. Similar invasions from the sea have been also postulated for Dinarides’ cave fauna (e.g. Sket, 2012), through the brackish/freshwater coastal sediments, especially during the Messinian Salinity Crisis (Boutin & Coineau, 2000). Contrary to common reports on the hydrobioid stygobiont species endemic to very restricted area—often just one cave—the hydrobiid cave snail Fontigens tartarea Hubricht, 1963 is known from dozens of caves in West Virginia (North America), its geographic range reaching 200 km and showing patchy distribution, sporadic occurrence, and high variability of the shell (Hershler & Holsinger, 1990; Culver, 2012). In caves, one can also find some widely distributed generalist species, also inhabiting surface waters—such as the pulmonate gastropod Ancylus fluviatilis O. F. Müller, 1774 (reported by e.g. Sket, 2012; it has also been found in several interstitial pumped materials during our research), Radomaniola Szarowska, 2006, and the isopod crustacean Asellus aquaticus (Linnaeus, 1758) (reported by: Verovnik et al., 2003; Verovnik, 2012). Molecular studies on the stygobiotic genus Bythiospeum Bourguignat, 1882 revealed the relatively late post-glacial colonisation of large regions north of the Alps by one genetically uniform species, while two other species of this genus had a very limited distribution colonised from their post-glacial refugia (Richling et al., 2016). This pattern also suggested that the river alluvium and gravels are effective ways of dispersal for some species of Bythiospeum, but which are not so effective for some other species, not inhabiting gravel alluvia, which are rare in the karst Balkan regions.
In general, the central question for understanding the biogeography of the subterranean animals, for years known mostly from caves, is that of the relative role of surface and subterranean dispersal (Holsinger, 2005). In the case of obligatory subterranean aquatic species assuming isolation in caves is not obvious. In fact, there are many subterranean habitats which are not caves, but which also harbour eyeless, depigmented animals (Culver & Pipan, 2009, 2014), especially in unconsolidated sediments bordering and underlying streams and rivers (hyporheic zone). They are parts of the interstitial habitat, neither rare nor discontinuous, thus, making possible migration between caves (Lamoreaux, 2004; Culver et al., 2009; Dole-Olivier, 2011). There are some studies confirming this possibility (e.g. Buhay & Crandall, 2005), but they are still few and are mostly devoted to the Crustacea (e.g. Lefébure et al., 2007; Eme et al., 2013), rather than the Gastropoda. The finds of live Paladilhiopsis oshanovae (Pintér, 1968) from the alluvial springs of the Danube (G. Majoros, pers. com.) clearly confirm alluvial gravel deposits as a habitat of this species; thus, the ways of its dispersal become obvious.
Pešić & Glöer (2013) described a new genus of the stygobiont snail—Montenegrospeum Pešić et Glöer, 2013, with its type species Bythiospeum bogici Pešić et Glöer, 2012 from the underground waters of the spring Taban, in the central part of Montenegro. Falniowski et al. (2014) applied molecular markers (COI and 18S) to resolve phylogenetic relationships of Montenegrospeum, as belonging to the Hydrobiidae, rather than the Moitessieriidae. Later, Grego et al. (2018) described another nominal species: M. sketi Grego et Glöer, 2018, whose description was based on the slight differences in the shell morphology and geographic distribution, but which was not confirmed by the molecular differences (Grego et al., 2018). The aim of the present paper is to check the genetic structure (applying mtDNA cytochrome oxidase subunit I (COI), three nuclear markers and RAPD analysis) of the populations of Montenegrospeum inhabiting diverse subterranean habitats, including caves and interstitial ones, to estimate the levels of gene flow, to confirm the hypothesis that these snails can migrate through them and forms a genetically uniform metapopulation through a relatively long distance as a result of a high level of gene flow.
Material and methods
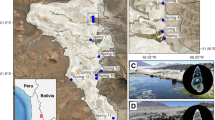
The snails were collected in 2018 and 2019, 27 specimens from 11 localities (Table 1), distributed in Croatia, Bosnia and Herzegovina, and Montenegro (Figs. 1 and 2). They were either collected by hand and sieve in caves and springs, or with a pump applying the Bou-Rouch technique (Bou & Rouch, 1967), to sample interstitial fauna below the bottom of streams, at a depth of about 50 cm. The tube was inserted in the streambed five times, and 20 L were pumped each time. Samples were sieved through a 500 μm sieve and fixed in 80% analytically pure ethanol, replaced twice, and later sorted. Next, the snails were put in fresh 80% analytically pure ethanol and kept at − 20 °C temperature in a refrigerator.
The shells were photographed with a Canon EOS 50D digital camera, under a Nikon SMZ18 microscope with dark field. Morphometric parameters of the shell (Table 2) were measured by one person using a Nikon DS-5 digital camera and ImageJ image analysis software (Rueden et al., 2017). The linear measurements were then logarithmically transformed; for angular measurements, the arcsine transformation was applied. Principal component analysis (PCA), based on the matrix of correlation, was computed, applying a descriptive, non-stochastic approach. The original observations were projected into PC space, to show relationships between the specimens, without any classification given a priori. For PCA analysis ClustVis 2.0 web tool (Metsalu & Vilo, 2015) was used.
DNA was extracted from whole specimens including their crushed shell; tissues were hydrated in TE buffer (3 × 10 min); then total genomic DNA was extracted with the SHERLOCK extraction kit (A&A Biotechnology), and the final product was dissolved in 20 μl of tris–EDTA (TE) buffer. The concentration of DNA was measured with EPPENDORF BIOPHOTOMETER. The extracted DNA was stored at − 80 °C at the Department of Malacology, Institute of Zoology and Biomedical Research, Jagiellonian University in Kraków (Poland).
A fragment of mitochondrial cytochrome oxidase subunit I (COI) and three nuclear fragments (18S ribosomal RNA—18S, 28S ribosomal RNA—28S, histone H3—H3) were sequenced, as commonly used markers in phylogenetic studies of Truncatelloidea. Some other markers (nuclear internal transcription spacers ITS-1 and ITS-2, elongation factor—EF as well as mitochondrial 16S ribosomal RNA—16S) did not amplify despite numerous attempts. Details of PCR conditions, primers used, and sequencing techniques were given in Szarowska et al. (2016a). The Sanger sequencing was performed in Genomed Company in Warsaw, Poland. Sequences were initially aligned in the MUSCLE (Edgar, 2004) programme in MEGA 7 (Kumar et al., 2016). The correctness of the alignment was checked in BIOEDIT 7.2.5 (Hall, 1999), this programme was also used to translate sequences and check for reading frame and stop codons. Uncorrected p-distances were calculated in MEGA 7. The estimation of the proportion of invariant sites and the saturation test (Xia, 2000; Xia et al., 2003) were performed using DAMBE (Xia, 2018). In the phylogenetic analysis, additional sequences from GenBank were used (Table 3). The phylogenetic analysis was performed applying two approaches: Bayesian Inference (BI) and Maximum Likelihood (ML). In the BI analysis, the GTR + I + Γ model of nucleotide substitution was applied. Model was selected using MRMODELTEST 2.3 (Nylander, 2004). The Bayesian analyses were run using MrBayes v. 3.2.3 (Ronquist et al., 2012) with defaults of most priors. Two simultaneous analyses were performed, each with 10,000,000 generations, with one cold chain and three heated chains, starting from random trees and sampling the trees every 1000 generations. The first 25% of the trees were discarded as burn-in. The analyses were summarised as a 50% majority-rule tree. Convergence was checked in Tracer v. 1.7 (Rambaut et al., 2018). FigTree v.1.4.4 (Rambaut, 2010) was used to visualise the trees. The Maximum Likelihood analysis was conducted in RAxML v. 8.2.12 (Stamatakis, 2014) using the “RAxML-HPC v.8” on XSEDE (8.2.12) tool via the CIPRES Science Gateway (Miller et al., 2010). We applied the GTR model whose parameters were estimated by RaxML (Stamatakis, 2014). Rapid bootstrap was calculated. Two species delimitation methods were performed: Poisson Tree Processes (PTP) (Zhang et al., 2013) and Automatic Barcode Gap Discovery (ABGD). The PTP approach was run using the web server https://species.h-its.org/ptp/, with 100,000 MCMC generations, 100 thinning, and 0.1 burn-in. We used RAxML output phylogenetic tree. The ABGD approach is using the web server (https://bioinfo.mnhn.fr/abi/public/abgd/abgdweb.html) and the default parameters. To infer haplotype network of the marker, we used a median-joining calculation implemented in NETWORK4 (Bandelt et al., 1999). The parameters describing a genetic variation and other statistics were calculated with DnaSP software (Librado & Rozas, 2009) and Arlequin v. 3.5 (Excoffier & Lischer, 2010). Arlequin was also used to perform Tajima’s D test of neutrality (Tajima, 1989a, 1989b, 1993). The geographical distances between the localities were calculated with Geographic Distance Matrix Generator v. 1.2.3 (Ersts, 2020). Mantel tests were performed with NTSYSpc (Rohlf, 1998).
In the RAPD analysis, run to obtain another source of information about genetic diversity, five primers were used (Table 4), based on the data from Kulsantiwong et al. (2013). The RAPD reaction mix contained: 10 μl of 1 × reaction buffer, 1 unit of Taq DNA polymerase (Thermo Fisher Scientific), 2.5 mM of potassium chloride, 0.2 μM of primer, 0.2 mM of dNTPs mix, and 10 ng of DNA0. The amplification conditions were as follows: the first step at 94 °C for 5 min, followed by 41 cycles of 1 min each at 94 °C, 1 min 30 s at 35 °C, and then 2 min at 72 °C. The final elongation step was for 5 min at 72 °C. The amplification products were separated in 2% agarose gel with ethidium bromide in TBE buffer. The banding patterns were visualised under UV light. The bands were scored visually, and those of similar molecular size (for the same primer) were assumed to be homologous. Each band was treated as an independent locus with two alleles: the presence or absence of the band. The PopGen 32 software package (Yeh & Yang, 2000) was used to estimate the genetic variation. The following parameters were calculated: mean number of alleles per locus, effective number of alleles per locus, percentage of polymorphic loci and Nei’s (1973) gene diversity, and the Shannon index. This same software was used to calculate the Nei’s genetic distances between populations.
Results
Both techniques of species delimitation resulted in distinction of three mOTUs. The shells of the sequenced specimens (Fig. 3A–Y) were slightly variable, only their size differed between the specimens. Neither the paratype of Montenegrospeum sketi (Fig. 3A), nor the specimens of molecularly distinct operational taxonomic unit (mOTU—see below) B and C differed morphologically from the other. Moreover, PCA analysis illustrates no variability and no morphological distinctiveness of the three clades as well as for populations of the Montenegrospeum (Fig. 4).
Shells of the sequenced specimens of Montenegrospeum: A-U—molecularly distinct operational unit (mOTU) A: A 2H70, locality 1, B 2D09, locality 2, C 2D10, locality 2, D 2D13, locality 2, E 1T57, locality 3, F 2D02, locality 4, G 2D04, locality 4, H 2J40, locality 5, I 2J41, locality 5, J 2J42, locality 5, K 2J37, locality 6, L 2J38, locality 6, M 2J39, locality 6, N 2C43, locality 8, O 2C44, locality 8, P 2C45, locality 8, Q 2C15, locality 9, R 1T19, locality 10, S 2C18, locality 10, T 2C40, locality 10, U 2C19, locality 10; V mOTU B, 2C28, locality 5; W–Y mOTU C: W 2C41, locality 7, X–Y locality 10 (2G20 and 2G23, respectively)
Principal component analysis (PCA) on the shell of Montenegrospeum based on the measured characters given in Table 2
The tests for COI and H3 revealed no saturation. We obtained 25 new sequences of each marker: COI (451 bp, GenBank Accession numbers OK001173-OK001197), 18S (401 bp, GenBank Accession numbers OK001198-OK001222), 28S (512 bp, GenBank Accession numbers OK001281-OK001305), and H3 (307 bp, GenBank Accession numbers OK033490-OK03351). For three nuclear fragments, all sequences were identical, so all analyses were made for the COI fragment. In the analysis of the COI, we included also all the sequences of Montenegrospeum available in the GenBank as well as for four sequences of the genera previously identified as the closest of Montenegrospeum for rooting the trees (Table 3).
We obtained 18 haplotypes for COI, with the haplotype diversity (Hd) 0.955 ± 0.024. In total, 44 sites were variable and nucleotide diversity per site (Pi) was 0.0276 ± 0.007.
The maximum likelihood (ML) phylogram (Fig. 5) presented the same topology as the consensus tree based on the Bayesian inference (BI). Reconstructing a shallow phylogeny of closely related taxa, it showed high values of bootstrap support or Bayesian probability, respectively. COI tree topology consisted of three molecular groups—mOTUs (confirmed by PTP and ABGD analysis), considering the identity of the sequences in the other loci they rather do not represent distinct species, although the values of pairwise p-distances for COI (Table 5) are within the interspecies level. All but four sequences of the snails, inhabiting ten localities (the type locality of M. sketi included: Fig. 1), formed mOTU A. Within this group, the genetic variation was very restricted, despite its wide (for stygobionts) geographic distribution, reaching 236 km (Table 5); three haplotypes were found in two (H9 and H15) or three (H11) populations (Table 1) mOTU B was found as a single specimen at one locality, and mOTU C as three specimens at two localities (Figs. 1 and 5). The above pattern of genetic differentiation was further supported by the results of AMOVA (Table 6). As much as 86.40% of the variance occurred between the mOTUs, confirming low genetic diversity within vs high between the mOTUs. Within the mOTUs, among the populations, only 7.36% of the variance was noted, and was not much more than 6.25% within the populations.
The results of Tajima’s D test of neutrality were statistically insignificant for the mOTU A. For the other two clades (B and C) they were statistically significant, their values were 0.0000, rejecting selection (although in the mOTU B, with one sequence, the value was meaningless). In the clade A, the result was nearly significant (p = 0.8890), and D = 1.1291. Tajima’s D > 0 reflects, in general, scarce rare alleles (in our case substitutions), balancing selection and sudden population contraction. With one polymorphic locus only, this result should be interpreted with caution. The results of Fu’s F test were statistically insignificant. The FST pairwise values between the mOTUs were statistically not significant.
The Mantel test, checking the association between the matrices of pairwise p-distance and geographic distance, thus, testing the isolation-by-distance model (Table 5, Fig. 6), with 9999 permutations, resulted in p [random Z ≤ observed Z] = 0.0318. The test between the matrices of geographic distance and FST (θ) (Table 7, Fig. 6), with 9999 permutations, resulted in p [random Z ≤ observed Z] = 0.3021. Pairwise theta (FST), Fisher’s estimator of the amount of structuring of a population into subpopulations, in general was high, but, with the exception of the ones between populations 2 and 4, as well as 10 and 4, not statistically significant. Nm, an estimator derived from Fisher’s F-statistics, may reflect the levels of gene flow, thus, migration in a subdivided population. In our populations, the values of Nm (Table 7) were, in general, high, with exceptions of the pairs of populations belonging to different mOTUs, or the pairs including the snails from the localities 10 and 5, inhabited by the representatives of two mOTUs, in sympatry.
The network of 18 found haplotypes (Fig. 7) confirms the distinctiveness of the mOTU C, but not necessarily that of the mOTU B. Within the most diversified mOTU A, there are 15 haplotypes, and the pattern more or less resembles the so-called “star phylogeny”.
In the RAPD analysis, each primer amplified from six to twelve fragments, with an average of eight, and among them, 38 bands (90%) were polymorphic (Table 4). Basic parameters of diversity are given in Table 8. Nei’s (1973) gene diversity ranged from 0.103 to 0.243 (mean 0.138, and the Shannon index ranged from 0.035 to 0.366, (mean 0.159)). The values were rather low, with the exception of the snails from the localities 7 and 10, inhabited each by two mOTUs. The Mantel test of association between the matrices of geographic distance and Nei’s genetic distances (Table 9, Fig. 8) with 9999 permutations, resulted in p [random Z ≤ observed Z] = 0.0189. There were not found to be any population-specific RAPD fragments, defined as RAPD bands found exclusively in one population in over 90% of individuals. Most of the detected RAPD bands were distributed across most populations (Fig. 9). In general, UPGMA clustering (Fig. 10) showed groupings of specimens in accordance with the localities from where they came, and within six main clusters. The first of them groups all individuals from the localities 1, 3, and 4. The second groups individuals from the locality 7, belonging to mOTU A. The third groups all the individuals from the localities 5 and 6, belonging to mOTU A. The fourth consists of specimens from the locality 2, while the fifth consists of all the individuals of mOTU C (however, high genetic distance within mOTU C may suggest the existence of two divergent groups). The sixth clusters the specimens from the localities 8, 9, 10 and a single individual of mOTU B from the locality 5 (Fig. 10).
In the karst areas of the Balkans the gravel substratum is not common, with interstitial waters usually filling small fractures in the rocks, making the collection of the meiofauna problematic, especially when using the Bou-Rouch pump. Coupled with low densities of such subterranean snails’ populations, most of the pumping does not result in the collection of live snails. We collected live specimens of Montenegrospeum at two localities: 2 and 10. Locality 2 was geographically most close (geographic distance 34 km) to the locality 4 (Table 4), which was a cave, and genetically, the populations were very close (p = 0.007; Table 4), confirming the possible migration from one locality to another. The locality 10 harboured two mOTUs: A and C. The mOTU C was found also in a spring at locality 7, which is 66 km far from the locality 10, and p = 0.066 (Table 4), and Nm equalled 0.000. For the representatives of the mOTU A, the localities closest to 10 were 9 (24 km), 11 (25 km), and 8 (35 km; Table 4), all being springs. The representatives of mOTU A from the locality 10 shared haplotypes with either locality 8 or 9.
Discussion
Neither morphology nor molecular data (Grego et al., 2018) confirmed the species distinctiveness of Montenegrospeum sketi. Thus, M. sketi becomes a younger synonym of B. bogici. Such unjustified species descriptions are not rare in truncatelloid gastropods (e. g. Wilke & Falniowski, 2001; Falniowski & Beran, 2015; Falniowski, 2018; Osikowski et al., 2018). Again, for the reasons outlined by e.g. Falniowski (1987, 2018), the morphostatic mode of evolution, as defined by Davis (1992), and common cryptic species, morphologically based species identification in the Truncatelloidea is often impossible. On the other hand, in our Montenegrospeum molecular data, we uncovered three molecularly distinct units; their distinctiveness is within the range of COI interspecies differentiation in the Truncatelloidea (e.g. Bichain et al., 2007; Falniowski & Szarowska, 2011), although not confirmed by the other loci, thus, not satisfying the criteria of species delimitation (Fišer et al., 2018). Perhaps mOTU C represents a distinct species, which is not reflected in its morphology—thus, there may be one more example of a cryptic species, not as rare as it had been thought before the common application of molecular techniques in taxonomy (e. g. Bickford et al., 2006; Pfenninger & Schwenk, 2007; Macher et al., 2016; Razkin et al., 2016, 2017). Cryptic species seem especially common in subterranean habitats (e.g. Culver & Pipan, 2009; Culver, 2012). In our case, the species distinctiveness of mOTU C is somewhat confirmed by the sympatric occurrence with mOTU A. Thus, instead of one stygobiont species, rather widely geographically dispersed, we may observe one widely distributed species (235 km of linear distance is neither characteristic nor common for subterranean species: e.g. Culver & Pipan 2009), and two possible cryptic species whose ranges are restricted to one or two localities known so far. Such a pattern has already been found for some stygobiont species (e.g. Eme et al., 2013; Gorički & Trontelj, 2006).
Within the mOTU A, which includes the topotypes of M. bogici and, thus, represents M. bogici s. stricto, the interpopulation differentiation is low, lower than in, for example, the crenobiont/stygophile (inhabiting springs and facultatively subterranean habitats) Bythinella Moquin-Tandon, 1856 (Falniowski et al., 1998, 2009; Falniowski & Szarowska, 2011; Osikowski et al., 2015; Szarowska et al., 2016b). Low values of p-distance and low levels of intrapopulation polymorphism may suggest rather high levels of gene flow, and either sudden population contraction (suggested by Tajima's test, although the results have been marginally significant only), or a “star phylogeny” pattern visible in the haplotype network, which could be interpreted as an expected signature for a species that has expanded its range rather recently, from a small or modest number of founders (Avise, 2000). As was stressed by Endler (1977), adaptive differences among populations can be maintained by natural selection even under high levels of gene flow, as well as selection may mimic effects of gene flow. This similarity may be also due to numerous stochastic factors, area effects, etc. Thus, another possible background of low variation may be selection, which is rather obvious in COI, a part of the respiratory chain. However, such selection should act uniformly in such a rather homogenous (at least concerning respiration: in a general low oxygenated) habitat as a subterranean one, but the mOTUs B and C, living in the same habitats (even in sympatry with A) are molecularly markedly distinct, which makes selection a much less probable factor shaping low molecular diversity. If Nm > 1, the allele frequencies in the subpopulations remain homogenised (Wright, 1931, 1969). If Nm < 1 but is still positive, an equilibrium based on the rate of mutation, migration, and genetic drift will be established. In our populations, the values of Nm were, in general, high, with exceptions of the pairs of populations belonging to different mOTUs, or the pairs including the snails from the localities 10 and 5, inhabited by the representatives of two mOTUs, in sympatry. The infinite-island model of interpopulation differentiation (Wright, 1978), expected for isolated populations, does not characterise the observed pattern. Together with the results of the Mantel test, it suggests isolation-by-distance (although slightly marked), rather than the stepping-stone model. RAPD confirms the same conclusion. Another three available, nuclear loci were not varied, which only confirms close relationships/high levels of gene flow of the studied individuals.
The low subsurface dispersal hypothesis (Lefébure et al., 2006), most popular for many years, states that after a vicariant event, invasion of subterranean habitat (group of caves) followed by extinction of the surface ancestor, there is little or no dispersal between the subterranean microhabitats. Inhabitants of subterranean habitats were often traditionally understood as being either living fossils, relic species (last survivors of ancient radiation), or relict species (geographically separated from related species) and showing no progressive evolution, being isolated an extremely space-limited habitat. This already abandoned concept seems to be the background of the popular practice among some taxonomists, many of them rather speleologists than biologists—to describe distinct species or at least subspecies for nearly each cave (two species described from two caves at the same village may serve as an example), with no understanding of either mechanisms of speciation or of potentially available dispersal pathways. Molecular markers, especially DNA partial sequences, have identified numerous cryptic subterranean species (e.g. Trontejl et al., 2009) but have also rejected the species distinctiveness of many nominal species described following the low subsurface dispersal hypothesis (e.g. Osikowski et al., 2017, 2018). However, Christman et al. (2005) in their study of the troglobiotic (terrestrial) fauna in eastern North America reported as many as 45% (211 of 467 species) to be single cave endemics.
Such high levels of endemism seem not so characteristic for the aquatic, stygobiont fauna. Ward & Palmer (1994) in their overview of dispersal of meiofauna, applicable to stygobionts in general, stressed the connections of subterranean habitats, such as caves and springs, by alluvial aquifers, with their sand and gravel deposited by flowing waters. Williams (2008) stressed that in almost all caves, the rock around is fractured, forming small solution tubes that allow subsurface connections between caves; in karst areas, epikarst and associated vertical downward percolation of water is more or less continuous. Their opinion was confirmed by phylogenetic analysis of four species of Proasellus Dudich, 1925 (belonging to the Crustacea: Isopoda) by Eme et al. (2013). There are some more studies confirming this possibility, which are mostly devoted to the Crustacea (e.g. Lefébure et al., 2007; Villacorta et al., 2008; Malard et al., 2009; Eme et al., 2013), but also to anchialine annelids (Gonzalez et al., 2017) which are the prevailing group in the stygobiont fauna (Sket, 2012). To our knowledge, there are no such studies on any stygobiont gastropod species. Our study, although based on a few specimens—as already stressed stygobiont gastropods do not form dense populations and are not easily collectable—presents the same picture: dispersal through the subterranean interstitial waters which they also inhabit, leading to low levels of isolation and the resultant endemism.
There may be various hydrological routes of migration of the stygobiont fauna. For example, in the Dinarids, there are large flat depressions (karst poljes), some with neither aerial inflow nor aerial outflow of water (closed karst poljes) and having insoluble polje-floor sediments. They are periodically flooded. During such a flood, aquatic cave animals are frequently washed out of their subterranean habitat, and some of them may reach caves at other parts of the polje, or even reach a subterranean connection to another polje. Such a route of migration was suggested by Zakšek et al. (2009) for the shrimp Troglocaris Dormitzer, 1853. However, Troglocaris is a swimmer, and for creeping snails, such a means of dispersal seems less probable, although some may possibly passively wash out and disperse.
The relatively wide geographical distribution of Montenegrospeum bogici, over 230 km, with one or two cryptic species at one/two populations, presents a pattern already found for some other, non-gastropod more motile stygobionts. The phylogenetic analysis of five morphospecies of the crustacean Proasellus, widely distributed (more than 200 km) along the hyporheic corridors of rivers, resulted in the discovery of ten new cryptic species, seven known from a single locality and being peripheral isolates (Eme et al., 2013)—such peripheral isolates may be mOTUs B and C. The most widely known and familiar stygobiont animal Proteus anguinus Laurenti, 1768, the European Cave Salamander, has a wide geographical range along the western part of former Yugoslavia, although the range is discontinuous (Sket, 1997). Gorički & Trontelj (2006) sequenced several regions on its mtDNA and distinguished six groups of its populations, with genetic divergence between them higher than between most species of salamanders within a genus. The greatest geographic extent of a single group does not extend 200 km, compared with 500 km for all the nominal species. All the distinguished six species were cryptic. The dispersal pattern of Montenegrospeum also strongly resembles the one described for Bythiospeum presented by Richling et al. (2016).
The relatively broad geographic range and high levels of gene flow in M. bogici observed over areas without alluvial connection need some possible explanation. The hydrogeological map of the species range (Fig. 11) suggests that M. bogici may inhabit not only the alluvial gravels and the spring heads, but should also be capable of spreading through the cave streams and lakes. Additionally, the species may use the intermittent connections between the aquifers of river drainage basins. Such an intermittent connection between the Drin (Zeta) and Trebišnjica River Basins is relatively well understood through Nikšičko Polje (Gornjepolski Vir) to Nikšičko Vrelo near Bileća (divergence an in Fig. 11). The species also passes the second intermittent divergence line between the Trebišnjica and Neretva River Basins, where it exists between the Dabarsko and Fatničko Poljes. Although this hydrological connection has been previously proven to exist when the karst groundwater exceeds 400 m a.s.l. elevation, the identical stygobiont malacofauna of both Poljes, with Plagigeyeria reischuetzorum Grego, 2020 and Travunijana gloeri Grego, 2020, now adds further evidence to confirm the existence of this connection. Furthermore, the stygofauna of both basins may be in contact through the springs in the delta of the Lower Neretva River in Hutovo Blato/Deranjsko Jezero and also near Metković (springs Bajovci, Sjekoše, Glušći, Bijeli Vir, Mlinišče). The third, hydrogeologically least understood, unremittent connection possibly exists between the Neretva and Cetina Basins. Most likely it is formed by occasional groundwater flow around Imotsko Polje, from Prološko Blato to the spring of Ričina in Buško Blato, or may be directly to the spring Ruda Beguša. The high levels of gene flow presented above indirectly prove the existence of this groundwater connection, at least intermittently. The finds of Montenegrospeum shells at the nearby spring Peć Mlini (Tihaljina, BiH) and in the spring Lukavac (Veliki Prolog, HR) indicate the wider distribution of the genus in areas between the hitherto-proven localities.
Hydrological map of the studied area with possible intermittent groundwater connections among Drin-Trebišnjica (A), Trebišnjica—Neretva (B), and Neretva—Cetina (C) River basins. Light blue arrows: Groundwater flow; Light blue ring: Karst spring; Blue ring with green centre and black X: Estavelles; dark green diamond: Submarine or brackish karst spring; Grey fields: permeable (conductive) karst; White field: not permeable (non-conductive) non-karstic bedrock; Yellow fields: Permeable (conductive) alluvial gravel; Orange fields: Alluvial sands with limited permeability (conductivity)
Concluding our results, in general, neither the infinite-island model of interpopulation differentiation, which would be expected for isolated populations, nor the stepping-stone one, but rather the isolation-by-distance model explained the observed pattern. Our results suggest that interstitial habitats provide ways of migration for the stygobiont M. bogici, as has been already suggested for other subterranean gastropods.
Data availability
Sequence data are also downloaded at GenBank; accession numbers xxxxx-xxxxx.
Code availability
Software application—BioEdit 7.2.5.
References
Avise, J. C., 2000. Phylogeography. The History and Formation of Species. Harvard University Press, Cambridge, MA.
Bandelt, H. J., P. Forster & A. Röhl, 1999. Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution 16: 37–48.
Beran, L., S. Hofman & A. Falniowski, 2015. Tanousia zrmanjae (Brusina, 1866) (Caenogastropoda: Truncatelloidea: Hydrobidae): a living fossil. Folia Malacologica 23: 263–271.
Bernasconi, R., 2000. Révision du genre Bythinella (Moquin-Tandon, 1855) (Gastropoda, Prosobranchia, Hydrobiidae, Amnicolinae, Bythinellini). Documents Malacologiques, hors série1: 1–126.
Bernasconi, R. & A. Riedel, 1994. Mollusca. In Juberthie, C. & V. Decu (eds), Encyclopedia Biospeleologica I. Société Biospéologie, Moulis: 54–61.
Bichain J. M., P. Gaubert, S. Samadi & M. C. Boisselierdubayle, 2007. A gleam in the dark: phylogenetic species delimitation in the confusing spring-snail genus Bythinella Moquin-Tandon, 1856 (Gastropoda: Rissooidea: Amnicolidae). Molecular Phylogenetics and Evolution 45: 927–941.
Bickford D., D. J. Lohman, N. S. Sodhi, P. K. L. Ng, R. Meier, K. Winker & I. Das, 2006. Cryptic species as a window on diversity and conservation. Trends in Ecology and Evolution 22: 148–155.
Boeters, H. D., 1979. Species concept of prosobranch freshwater molluscs in Western Europe, 1. Malacologia 18: 57–60.
Bole, J. & F. Velkovrh, 1986. Mollusca from continental subterranean aquatic habitats. In Botosaneanu, L. (ed.), Stygofauna Mundi. E. J. Brill, Leiden: 177–206.
Bou, C. & R. Rouch, 1967. Un nouveau champ de recherches sur la faune aquatique souterraine. Comptes Rendus de’lAcadémie des Sciences Series III Sciences 265: 369–370.
Boutin, C. & N. Coineau, 2000. Evolutionary rates and phylogenetic age in some stygobiontic species. In Wilkens, H., D. C. Culver & W. F. Humphreys (eds), Subterranean Ecosystems. Elsevier Press, Amsterdam: 433–451.
Buhay, J. E. & K. E. Crandall, 2005. Subterranean phylogeography of freshwater crayfishes shows extensive gene flow and surprisingly large population sizes. Molecular Ecology 14: 4259–73.
Christman, M. C., D. C. Culver, M. Madden & D. White, 2005. Patterns of endemism of the eastern North American cave fauna. Journal of Biogeography 32: 1441–1452.
Culver, D. C., 2012. Mollusks. In White, W. B. & D. C. Culver (eds), Encyclopedia of Caves (Second Edition). Academic Press, Amsterdam: 512–517.
Culver, D. C. & B. Sket, 2000. Hotspots of subterranean biodiversity in caves and wells. Journal of Cave and Karst Studies 62: 11–17.
Culver, D. C. & T. Pipan, 2009. The Biology of Caves and Other Subterranean Habitats. Oxford University Press, Oxford.
Culver, D. C. & T. Pipan, 2014. Shallow Subterranean Habitats. Ecology, Evolution and Conservation. Oxford University Press, Oxford.
Culver, D. C., T. Pipan & K. Schneider, 2009. Vicariance, dispersal and scale in the aquatic subterranean fauna of karst regions. Freshwater Biology 54: 918–929.
Davis, G. M., 1992. Evolution of prosobranch snails transmitting Asian Schistosoma: coevolution with Schistosoma: a review. Progress in Clinical Parasitology 3: 145–204.
Dole-Olivier, M. J., 2011. The hyporheic refuge hypothesis reconsidered: a review of hydrological aspects. Marine and Freshwater Research 62: 1281–1302.
Edgar, R. C., 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32: 1792–1797.
Eme, D., F. Malard, L. Konecny-Dupré, T. Lefébure & C. J. Douady, 2013. Bayesian phylogeographic inferences reveal contrasted colonization dynamics among European groundwater isopods. Molecular Ecology 22: 5685–5699.
Endler, J., 1977. Geographic Variation, Speciation, and Clines. Princeton University Press, Princeton, NJ.
Ersts, P. J., 2020. Geographic Distance Matrix Generator (version 1.2.3). American Museum of Natural History, Center for Biodiversity and Conservation. http://biodiversityinformatics.amnh.org/open_source/gdmg.
Excoffier, L. & H. E. Lischer, 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources 10: 564–567.
Falniowski, A., 1987. Hydrobioidea of Poland (Prosobranchia: Gastropoda). Folia Malacologica 1: 1–122.
Falniowski, A., 2018. Species Distinction and Speciation in Hydrobioid Gastropods (Mollusca: Caenogastropoda: Truncatelloidea). Journal of Archives of Zoological Studies 1: 1–6.
Falniowski, A. & M. Szarowska, 2011. Radiation and phylogeography in a spring snail Bythinella (Mollusca: Gastropoda: Rissooidea) in continental Greece. Annales Zoologici Fennici 48: 67–90.
Falniowski, A. & M. Szarowska, 2013. Phylogenetic relationships of Dalmatinella fluviatilis Radoman, 1973 (Caenogastropoda: Rissooidea). Folia Malacologica 21: 1–7.
Falniowski, A. & L. Beran, 2015. Belgrandiella A. J. Wagner, 1928 (Caenogastropoda: Truncatelloidea: Hydrobiidae): how many endemics? Folia Malacologica 23: 187–191.
Falniowski, A. & S. Sarbu, 2015. Two new Truncatelloidea species from Melissotrypa Cave in Greece (Caenogastropoda). ZooKeys 530: 1–14.
Falniowski, A., M. Szarowska, W. Fiałkowski & K. Mazan, 1998. Unusual geographic pattern of interpopulation variation in a spring snail Bythinella (Gastropoda: Prosobranchia). Journal of Natural History 32: 605–616.
Falniowski, A., K. Mazan & M. Szarowska, 1999. Homozygote excess and gene flow in the spring snail Bythinella (Gastropoda: Prosobranchia). Journal of the Zoological Systematics and Evolutionary Research 37: 165–175.
Falniowski, A., M. Szarowska, I. Sirbu, A. Hillebrand & M. Baciu, 2008. Heleobia dobrogica (Grossu & Negrea, 1989) (Gastropoda: Rissooidea: Cochliopidae) and the estimated time of its isolation in a continental analogue of hydrothermal vents. Molluscan Research 28: 165–170.
Falniowski, A., M. Szarowska & I. Sirbu, 2009. Bythinella Moquin-Tandon, 1856 (Gastropoda: Rissooidea: Bythinellidae) in Romania: species richness in a glacial refugium. Journal of Natural History 43: 2955–2973.
Falniowski, A., V. Pešić & P. Glöer, 2014. Montenegrospeum Pešić et Glöer, 2013: a representative of Moitessieriidae? Folia Malacologica 22: 263–268.
Fišer, C., C. T. Robinson & F. Malard, 2018. Cryptic species as a window into the paradigm shift of the species concept. Molecular Ecology 27: 613–635.
Fretter, V. & A. Graham, 1994. British Prosobranch Molluscs. Their Functional Anatomy and Ecology. Revised and Updated Edition. The Ray Society, London.
Georgiev, D., 2013, Catalogue of the stygobiotic and troglophilous freshwater snails (Gastropoda: Rissooidea: Hydrobiidae) of Bulgaria with descriptions of five new species. Ruthenica 23: 59–67.
Giusti, F. & E. Pezzoli, 1982. Notes on the small Hydrobioidea in Italian subterranean waters: catalogue, biogeography and some systematic problems. Malacologia 22: 463–468.
Gladstone, N. S., M. L. Niemiller, B. Hutchins, B. Schwartz, A. Czaja, M. E. Slay & N. V. Whelan, 2021. Subterranean freshwater gastropod biodiversity and conservation in the United States and Mexico. Conservation Biology. https://doi.org/10.1111/cobi.13722
Glöer, P., 2002. Die Süswassergastropoden Nord- und Mitteleuropas. Bestimmungsschlussel, Lebensweise, Verbreitung. Die Tierwelt Deutschlands. Teil 73. Conchbooks, Hackenheim.
Glöer, P., J. Grego, Z. P. Eröss & Z. Fehér, 2015. New records of subterranean and spring molluscs (Gastropoda: Hydrobiidae) from Montenegro and Albania with the description of five new species. Ecologia Montenegrina 4: 70–82.
Gonzalez, B. C., A. Martínez, E. Borda, T. M. Iliffe, D. Fontaneto & K. Worsaae, 2017. Genetic spatial structure of an anchialine cave annelid indicates connectivity within-but not between-islands of the Great Bahama Bank. Molecular Phylogenetics and Evolution 109: 259–270.
Gorički, Š. & P. Trontelj, 2006. Structure and evolution of the mitochondrial control region and flanking sequences in the European cave salamander Proteus anguinus. Gene 378: 31–41.
Grego, J., P. Glöer, Z. P. Eröss & Z. Fehér, 2017. Six new subterranean freshwater gastropod species from northern Albania and some new records from Albania and Kosovo (Mollusca, Gastropoda, Moitessieriidae and Hydrobiidae) Subterranean Biology 23: 85–107.
Grego, J., P. Glöer, A. Falniowski, S. Hofman & A. Osikowski. 2019. New subterranean freshwater gastropods species from Montenegro (Mollusca Gastropoda Moitessieriidae and Hydrobiidae). Ecologica Montenegrina. https://doi.org/10.37828/em.2019.20.6.
Grego, J., P. Glöer, A. Rysiewska, S. Hofman & A. Falniowski, 2018. A new Montenegrospeum species from south Croatia (Mollusca: Gastropoda: Hydrobiidae). Folia Malacologica 26: 25–34.
Haase, M., 1995. The stygobiont genus Bythiospaeum in Austria: a basic revision and anatomical description of B. cf. geyeri from Vienna (Caenogastropoda: Hydrobiidae). Bulletin of American Malacological Union 11: 123–137.
Hall, T. A., 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98.
Hershler, R. & J. R. Holsinger, 1990. Zoogeography of North American hydrobiid snails. Stygologia 5: 5–16.
Hobbs, H. H. III, 2012. Diversity patterns in the United States. In White, W. B. & D. C. Culver (eds), Encyclopedia of Caves (Second Edition). Academic Press, Amsterdam: 251–264.
Hofman, S., A. Osikowski, A. Rysiewska, J. Grego, P. Glöer, D. Dmitrović & A. Falniowski, 2019a. Sarajana Radoman, 1975 (Caenogastropoda: Truncatelloidea): premature invalidation of a genus. Journal of Conchology 43: 407–418.
Holsinger, J. R., 2005. Vicariance and dispersalist biogeography. In Culver, D. C. & W. B. White (eds), Encyclopaedia of caves. Elsevier/Academic Press, Amsterdam: 591–599
Kulsantiwong, J., S. Prasopdee, S. Piratae, P. Khampoosa, A. Suwannatrai, W. Duangprompo, T. Boonmars, W. Ruangjirachuporn, J. Ruangsittichai, V. Viyanant, P. D. N. Hebert & S. Tesana, 2013. Species-specific primers designed from RAPD products for Bithynia funiculata, the first intermediate host of liver fluke, Opisthorchis viverrini, in North Thailand. Journal of Parasitology 99: 433–437.
Kumar, S., G. Stecher & K. Tamura, 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33: 1870–1874.
Lamoreaux, J., 2004. Stygobites are more wide-ranging than troglobites. Journal of Cave and Karst Studies 66: 18–19.
Lefébure, T., C. J. Douady, M. Gouy & J. Gibert, 2006. Relationship between morphology, taxonomy, and molecular divergence with Crustacea: proposal of a molecular threshold to help species definition. Molecular Phylogeny and Evolution 40: 435–447.
Lefébure, T., C. J. Douady, F. Malard & J. Gibert, 2007. Testing dispersal and cryptic diversity in a widely distributed groundwater amphipod (Niphargus rhenorhodanensis). Molecular Phylogenetics and Evolution 42: 676–686.
Librado, P. & J. Rozas, 2009. DnaSP v5 a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452.
Macher, J. N., M. Weiss, A. J. Beermann & F. Leese, 2016. Cryptic diversity and population structure at small scales: the freshwater snail Ancylus (Planorbidae, Pulmonata) in the Montseny mountain range. Annales de Limnologie - International Journal of Limnology 52: 387–399.
Malard, F., C. Boutin, A. L. Camacho, D. Ferreira, G. Michel, B. Sket & F. Stoch, 2009. Diversity patterns of stygobiotic crustaceans across multiple spatial scales in Europe. Freshwater Biology 54: 756–776.
Mammola, S., Lunghi, E., Bilandžija, H., Cardoso, P., Grimm, V., Schmidt, S. I., Hesselberg, T. & A. Martínez, 2021. Collecting eco-evolutionary data in the dark: Impediments to subterranean research and how to overcome them. Ecology and Evolution 11: 5911–5926.
Metsalu, T. & J. Vilo, 2015. Clustvis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Research, 43: 566–570.
Miller, M. A., W. Pfeiffer & T. Schwartz, 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the Gateway Computing Environments Workshop (GCE), 14 Nov., New Orleans, LA: 1–8.
Nei, M., 1973. Analysis of gene diversity in subdivided populations. Proceedings of the National Academy of Sciences of the United States of America 70: 3321–3323.
Nylander, J. A. A., 2004. MrModeltest v.2. Program distributed by the author. Uppsala: Evolutionary Biology Centre, Uppsala University.
Osikowski, A., D. Georgiev, S. Hofman & A. Falniowski, 2015. Does the genetic structure of spring snail Bythinella (Caenogastropoda, Truncatelloidea) in Bulgaria reflect geological history? ZooKeys 518: 67–86.
Osikowski, A., S. Hofman, D. Georgiev, A. Rysiewska & A. Falniowski, 2017. Unique, Ancient Stygobiont Clade of Hydrobiidae (Truncatelloidea) in Bulgaria: the Origin of Cave Fauna. Folia Biologica (Kraków) 65: 79–93.
Osikowski, A., S. Hofman, A. Rysiewska, B. Sket, S. Prevorčnik & A. Falniowski, 2018. A case of biodiversity overestimation in the Balkan Belgrandiella A. J. Wagner, 1927 (Caenogastropoda: Hydrobiidae): molecular divergence not paralleled by high morphological variation. Journal of Natural History 52: 323–344.
Pešić, V. & P. Glöer, 2012. A new species of Bythiospeum Bourguignat, 1882 (Hydrobiidae, Gastropoda) from Montenegro. Biologica Nyssana 3: 17–20.
Pešić, V. & P. Glöer, 2013. Montenegrospeum, a new genus of hydrobiid snails (Gastropoda: Risooidea) from Montenegro. Acta Zoologica Bulgarica 65: 565–566.
Pfenninger, M. & K. Schwenk, 2007. Cryptic animal species are homogeneously distributed among taxa and biogeographical regions. BMC Evolutionary Biology 7: 121.
Poulson, T. L., 2012. Food sources. In White W.B. & D. C. Culver (eds), Encyclopedia of Caves (Second Edition). Academic Press, Amsterdam: 323–334.
Purchon, R. D., 1977. The Biology of the Mollusca. International Series of Monographs in Pure and Applied Biology, Division: Zoology: 57. Pergamon Press, Oxford-New York-Toronto-Sydney-Paris-Frankfurt.
Rambaut, A., 2010. FigTree v1.3.1. http://tree.bio.ed.ac.uk/software/figtree.
Rambaut, A., Drummond, A. J., Xie, D., Baele, G., Suchard, M. A., & Susko, E. (2018). Posterior summarization in Bayesian phylogenetics using tracer 1.7. Systematic Biology, 67(5), 901–904.
Razkin, O., G. Sonet, K. Breugelmans, J. M. Madeira, B. J. Gómez-Moliner & T. Backeljau, 2016. Species limits, interspecific hybridization and phylogeny in the cryptic land snail complex Pyramidula: The power of RADseq data. Molecular Phylogenetics and Evolution 101: 267–278.
Razkin, O., B. Gomez-Moliner, K. Vardinoyannis, A. Martinez-Orti & M. J. Madeira, 2017. Species delimitation for cryptic species complexes: case study of Pyramidula (Gastropoda, Pulmonata). Zoologica Scripta 46: 55–72.
Reischütz, A., P. L. Reischütz & W. Fischer, 2008: Helleniká pantoía, 22. Zur Verbreitung der Gattung Bythinella Moquin-Tandon, 1856 (Gastropoda: Prosobranchia: Hydrobiidae) auf der Peloponnes (Griechenland). Nachrichtenblatt der Ersten Vorarlberger Malakologischen Gesellschaft 15: 35–36.
Richling, I., Y. Malkowsky, Y. Kuhn, H.-J. Niederhöfer & H. D. Boeters, 2016. A vanishing hotspot – impact of molecular insights on the diversity of Central European Bythiospeum Bourguignat, 1882 (Mollusca: Gastropoda: Truncatelloidea). Organisms Diversity and Evolution 17: 67–85.
Rohlf, F. J., 1998. NTSYSpc, numerical taxonomy and multivariate analysis system. Version 2.0. Exeter Software, Setauket, NY.
Ronquist, F., M. Teslenko, M. P. Vander, D. L. Ayres, A. Darling, S. Höhna & J. P. Huelsenbeck, 2012. Mr. Bayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542.
Rueden, D. T., J. Schindelin, M. C. Hiner, B. E. Dezonia, A. E. Walter, E. T. Arena & K. W. Eliceiri, 2017. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 18: e529.
Rysiewska, A., D. Georgiev, A. Osikowski, S. Hofman & A. Falniowski, 2016. Pontobelgrandiella Radoman, 1973 (Caenogastropoda: Hydrobiidae): A recent invader of subterranean waters? Journal of Conchology 42: 1–11.
Sbordoni, V., G. Allegrucci & D. Cesaroni, 2012. Population structure. In Culver, D. C. & W. B. White (eds), Encyclopedia of Caves (Second Edition). Academic Press, Amsterdam: 608–618.
Sket, B., 1997. Distribution of Proteus (Amphibia: Urodela: Proteidae) and its possible explanation. Journal of Biogeography 24: 263–280.
Sket, B., 2012. Diversity patterns in the Dinaric karst. In Culver, D. C. & W. B. White (eds), Encyclopedia of Caves (Second Edition). Academic Press, Amsterdam: 228–234.
Sket, B., K. Paragamian & P. Trontelj, 2004. A Census of the obligate subterranean fauna of the Balkan Peninsula. In Griffiths, H. I., B. Kryštufek & J. M. Reed (eds), Balkan Biodiversity: Pattern and Process in the European Hotspot. Kluwer Academic Publishers, Dodrecht: 309–322.
Stamatakis, A., 2014. RAxML Version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313.
Szarowska, M., A. Osikowski, S. Hofman & A. Falniowski, 2016a. Pseudamnicola Paulucci, 1878 (Caenogastropoda: Truncatelloidea) from the Aegean Islands: a long or short story? Organisms Diversity and Evolution 16: 121–139.
Szarowska, M., A. Osikowski, S. Hofman & A. Falniowski, 2016b. Do diversity patterns of the spring-inhabiting snail Bythinella (Gastropoda, Bythinellidae) on the Aegean Islands reflect geological history? Hydrobiologia 765: 225–243.
Tajima, F., 1989a. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595.
Tajima, F., 1989b. The effect of change in population size on DNA polymorphism. Genetics 123: 597–601.
Tajima, F., 1993. Measurement of DNA polymorphism. In: Takahata, N., Clark, A.G. (Eds.), Mechanisms of Molecular Evolution. Introduction to Molecular Paleopopulation Biology. Japan Scientific Societies Press, Sinauer Associates, Inc., pp. 37–59.
Trajano, E. & M. Cobolli, 2012. Evolution of lineages. In Culver, D. C. & W. B. White (eds), Encyclopedia of Caves (Second Edition). Academic Press, Amsterdam: 295–304.
Trontelj, P., C. J. Douady, C. Fišer, J. Gibert, Š Gorički, C. Lefébure, B. Sket & V. Zakšek, 2009. A molecular test for cryptic diversity in groundwater: how large are the ranges of macro-stygobionts? Freshwater Biology 54: 727–744.
Trueman, E. R., 1983. Locomotion in Molluscs. In Saleuddin, A. S. M. & K. M. Wilbur (eds), The Mollusca, Volume 4: Physiology, Part 1. Academic Press, Inc., (New York-London-Paris-San Diego-San Francisco-Sao Paulo-Sydney-Tokyo-Toronto): 155–198.
Verovnik, R., 2012. Asellus aquaticus: a model system for historical biogeography. In Culver D. C. & W. B. White (eds), Encyclopedia of Caves (Second Edition). Academic Press, Amsterdam: 30–36.
Verovnik, R., B. Sket, S. Prevorčnik & P. Trontelj, 2003. Random amplified polymorphic DNA diversity among surface and subterranean populations of Asellus aquaticus (Crustacea: Isopoda). Genetica 119: 155–165.
Ward, J. V. & M. A. Palmer, 1994. Distribution patterns of interstitial freshwater meiofauna over a range of spatial scales, with emphasis on alluvial river-aquifer systems. Hydrobiologia 287: 147–156.
Wilke, T., & A. Falniowski, 2001. The genus Adriohydrobia (Hydrobiidae: Gastropoda): polytypic species or polymorphic populations? Journal of Zoological Systematics and Evolutionary Research 39: 227–234.
Wilke, T., G. M. Davis, A. Falniowski, F. Giusti, M. Bodon & M. Szarowska, 2001. Molecular systematics of Hydrobiidae (Mollusca: Gastropoda: Rissooidea): testing monophyly and phylogenetic relationships. Proceedings of the Academy of Natural Sciences of Philadelphia 151: 1–21.
Williams, P. W., 2008. The role of the epikarst in karst and cave hydrogeology: a review. International Journal of Speleology 37: 1–10.
Villacorta, C., D. Jaume, P. Oromi & C. Juan, 2008. Phylogeography and evolution of the cave-dwelling Palmorhestia hypogea (Amphipoda: Crustacea) at La Palma (Canary Islands). BMC Biology 6: 7.
Yeh, F. C. & R. Yang, 2000. POPGENE Version 1.32. Dept. of Renewable Resources. Univ. of Alberta. https://sites.ualberta.ca/~fyeh/popgene.html
Wright, S., 1931. Evolution in Mendelian populations. Genetics 16: 97–159.
Wright, S., 1969. Evolution and the Genetics of Populations, Vol. 2, The Theory of Gene Frequencies. University of Chicago Press, Chicago.
Wright, S., 1978. Evolution and the genetics of populations. Vol. 4, Variability within and among natural populations. University of Chicago Press, Chicago.
Xia, X., 2000. Data analysis in molecular biology and evolution. Kluwer Academic Publishers, Boston.
Xia, X., 2018. DAMBE7: New and Improved Tools for Data Analysis in Molecular Biology and Evolution. Molecular Biology and Evolution 35: 1550–1552.
Xia, X., Z. Xie, M. Salemi, L. Chen & Y. Wang, 2003. An index of substitution saturation and its application. Molecular Phylogenetics and Evolution 26: 1–7.
Zakšek, V., B. Sket, S. Gottstein, D. Franjević & P. Trontelj, 2009. The limits of cryptic diversity in groundwater: Phylogeography of the cave shrimp Troglocaris anophthalmus (Crustacea: Decapoda: Atyidae). Molecular Ecology 18: 931–946.
Zhang, J., P. Kapli, P. Pavlidis & A. Stamatakis, 2013. A General Species Delimitation Method with Applications to Phylogenetic Placements. Bioinformatics 29: 2869–2876.
Acknowledgements
The study was supported by a grant from the National Science Centre 2017/25/B/NZ8/01372 to Andrzej Falniowski. We would like also to thank Dr Alejandro Martínez and an anonymous reviewer of the earlier version of the manuscript for their valuable comments and suggestions.
Funding
The study was supported by a grant from the National Science Centre 2017/25/B/NZ8/01372.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Handling editor: Diego Fontaneto
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Falniowski, A., Pešić, V., Lewarne, B. et al. Isolation and endemism in subterranean aquatic snails: unexpected case of Montenegrospeum bogici (Pešić et Glöer, 2012) (Gastropoda: Truncatelloidea: Hydrobiidae). Hydrobiologia 848, 4967–4990 (2021). https://doi.org/10.1007/s10750-021-04688-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-021-04688-6