Abstract
Ear length is an important component of maize grain yield. However, the ear length is a complex quantitative trait, and the underlying molecular mechanisms remain poorly understood. Here, the chromosome segment substitution line (CSSL) 1283 displayed a longer ear length compared with the recipient parent Xu178. An RNA sequencing analysis of Xu178 and CSSL1283 ears during three undifferentiated ear developmental stages identified 1,991 differentially expressed genes (DEGs). A gene ontology analysis of the DEGs showed that genes related to transcription factors and response to abiotic stimulus were significantly enriched. Furthermore, the expression of DEGs associated with AP2/EREBP and WRKY transcription factors and heat shock proteins was upregulated in CSSL1283. In addition, several genes encoding protein kinase were differentially expressed between Xu178 and CSSL1283. Our study provided a genetic resource for the dissection of the molecular mechanisms of ear-length development and for uncovering candidate genes to increase maize ear length.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Maize (Zea mays L.) is an important resource for food, feed and biofuel materials (Godfray et al. 2010). As the population rapidly increases, so will the demand for maize grain. Maize grain yield is determined by planting density, kernel weight and kernel number per ear (Huo et al. 2016). Ear length is closely associated with kernel number per ear, which ultimately affects the grain yield, therefore, it is a key target trait in high-yield maize breeding (Liu et al. 2012; Zhou et al. 2015).
Several quantitative trait loci (QTLs) for maize ear length have been identified (Huo et al. 2016; Ma et al. 2007; Vollbrecht and Schmidt 2009; Xiao et al. 2016; Yang et al. 2014; Zhou et al. 2015). However, only a few major QTLs, including KERNEL NUMBER PER ROW6 (KNR6) (Jia et al. 2020), KERNEL EAR LENGTH7 (qEL7) (Ning et al. 2021), YIGE1 (Luo et al. 2022) and EAR APICAL DEGENERATION1 (EAD1) (Pei et al. 2022), have been cloned. KNR6 encodes a serine/threonine-protein kinase and affects ear length and kernel number per row by interacting with an Arf GTPase-activating protein (Jia et al. 2020). qEL7 encodes an ethylene biosynthesis gene, ZmACO2. Variation in the promoter region of qEL7 leads to a decrease in the ethylene content of the ear, which promotes meristem development (Ning et al. 2021). YIGE1 encodes an unknown protein that may be involved in sugar and auxin signaling pathways, affecting inflorescence meristem (IM) activity and floret production in maize inflorescence morphogenesis (Luo et al. 2022). EAD1 encodes a cytoplasmic membrane-localized aluminum-activated malate transporter protein, which is specifically expressed in xylem conduit tissues of young maize ears. EAD1 regulates maize ear development by mediating transport in the vascular tissue of the young ears (Pei et al. 2022). Although significant progress has been made in understanding the regulation of ear length, knowledge of genes and biological pathways involved in ear-length development remains limited.
Maize ears start from the axillary meristems (AMs) in the axils of the leaves. In early ear development, switching from vegetative to reproductive development turns AMs into ear IMs. The IMs then elongate and produce spikelet-pair meristems (SPMs). Each SPM develops into two spikelet meristems (SMs), which become floral meristems that form kernels after fertilization (Eveland et al. 2013; Liu et al. 2015; Vollbrecht and Schmidt 2009). The initiation and maintained activity of these meristems determine the grain yield-related traits in maize. For instance, the kernel row number and grain yield are enhanced in the elite alleles of FASCIATED EAR2 (FEA2), FASCIATED EAR 3 (FEA3), UNBRANED3 (UB3) and KRN4, which are involved in the development of IMs and the number of SPMs (Bommert et al. 2013b; Chuck et al. 2014; Je et al. 2016; Liu et al. 2015).
Transcriptome profiling studies can explore spatial and temporal gene expression patterns. Thus, this method has been used to identify important genes and biological pathways responsible for tissue and organ development (Zhao et al. 2020). In maize, Yi et al. performed RNA sequencing (RNA-seq) and detected 1,093 genes, including 110 transcription factors (TFs) specifically expressed in seed (Yi et al. 2019). Notably, with a transcriptome dataset and gene overexpression lines, an AP2 TF BABY BOOM2 was shown to promote maize callus formation and transformation (Du et al. 2019). In addition, a transcriptomic comparison of maize non-stressed and heat-stressed pollen indicated that starch, lipid and energy biosynthetic processes are crucial for pollen development at the tetrad stage after transient heat stress (Begcy et al. 2019). The transcriptomic analysis revealed that some TFs correlated to drought stress and abiotic stress responses were enriched in the overexpression of AtGA2ox1 in maize (Chen et al. 2019).
Here, CSSL1283 displayed significantly longer ears and a greater kernel number per row compared with the recipient parent, Xu178. To reveal the underlying molecular mechanisms, we report the transcriptome profiling of CSSL1283 and its recipient parent, Xu178, at three early ear developmental stages. We identified the enriched processes and important genes involved in early ear-length development. This study promotes the understanding of the molecular mechanisms underlying ear-length development and will aid in the genetic improvement of maize ear length.
Results
Phenotypical analysis of CSSL1283
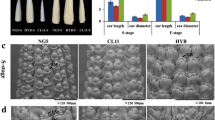
In total, 150 CSSLs were constructed using the inbred line Zong3 as the donor parent and Xu178 as the recipient parent (Mao et al. 2013). One of the lines, CSSL1283, displayed significantly longer mature ears and a higher kernel number per row compared with the recipient parent, Xu178 (Fig. 1A, J, K).
Phenotypic comparison between Xu178 and CSSL1283. A The mature ears of Xu178 and CSSL1283. Bar = 1 cm. B-I The ear primordia of Xu178 (B, D, F, H) and CSSL1283 (C, E, G, I) from stg1 (B and C), stg 2 (D and E), stg3 (F and G) and stg4 (H and I). Bars = 100 μm in B to E, 500 μm in F to I. J and K The comparison of ear length (J) and kernel number per row (K) at maturity ear. L The length of ear inflorescence meristem. Datas are presented as means ± SD, p-value is estimated by the two-tailed t-test, *P < 0.05, **P < 0.01
To gain a better insight into ear development, we tracked the growth of young Xu178 and CSSL1283 ears and measured the ear lengths at four different stages (stg 1-stg 4). Stg1 and stg2 ears initiated IMs and SPMs, respectively, with SMs formed at stg3 and floral meristems formed by stg4. The CSSL1283 ears were significantly longer than those of Xu178 at all four stages (Fig. 1B-I, L), indicating that the increased ear length of CSSL1283 occurs in an early stage of ear-length development.
Global transcript profiles of undifferentiated ear inflorescences
The phenotypic difference of CSSL1283 revealed that the expression of genes in early ear-length development contributed to the increased ear length. To explore regulatory networks of ear length in CSSL1283, we performed RNA-seq on Xu178 and CSSL1283 ears from stg1 to stg3. In total, 410.93 million high-quality reads were generated (Table 1) and then mapped to the maize B73 reference genome (RefGen_V4). On average, 87.89% of the total clean reads were uniquely mapped (Table 1). Gene expression levels were calculated as FPKM (fragments per kb exon model per million mapped fragments) values. Determinations of the Pearson correlation coefficients indicated that the expression levels between biological replicates were highly correlated (Table S1). We next compared the number of expressed genes for different samples. Genes with FPKM ≥ 1 were considered expressed genes. A total of 21,518 genes were expressed in at least one sample. Among these genes, 20,842 and 20,997 genes were detected in Xu178 and CSSL1283, respectively. In total, 94.44% (20,321/21,518) of the genes were detected in both Xu178 and CSSL1283 (Fig. 2A).
Analysis of differentially expressed genes. A Venn diagram of genes expressed in three stages of Xu178 and CSSL1283. The overlap indicates the number of genes commonly expressed in the two lines. B and C Venn diagrams of DEGs in three stages of Xu178 and CSSL1283. Up and down indicate that DEGs are upregulated and downregulated in Xu178 compared with CSSL1283, respectively. The overlaps indicate the numbers of DEGs shared in the upregulated and downregulated genes (B) and the three stages (C). D-F GO classifications of DEGs in stg1 (D), stg2 (E) and stg3 (F). F, P and C indicate the molecular function, biological process and cellular component, respectively. The top five most significantly enriched GO terms based on the FDR values are listed
Nine DEGs were selected for verification by qRT-PCR. The correlation coefficient of the qRT-PCR results and RNA-seq data was 0.85, indicating the RNA-seq data were reliable (Fig. 3).
Relative expression levels of nine genes as assessed by qRT-PCR. The histogram represents qRT-PCR data and the broken line represents RNA-seq data. Gray and black histograms indicate the Xu178 and CSSL1283 ears, respectively. Broken lines with hollow points and solid points indicate the Xu178 and CSSL1283 ears, respectively. Datas are presented as means ± SD, p-value is estimated by the two-tailed t-test, *P < 0.05, **P < 0.01
An overview of the differentially expressed genes (DEGs)
To identify the factors involved in the increased ear length of CSSL1283, we analyzed the DEGs between Xu178 and CSSL1283. Compared with CSSL1283, a total of 1,991 DEGs were identified in Xu178 using the cutoff criteria (FDR ≤ 0.05 and fold change ≥ 2), including 1,178 upregulated genes and 904 downregulated genes (Fig. 2B). In total, 908, 644 and 1,129 genes were differentially expressed in stg1, stg2 and stg3, respectively, suggesting more complicated regulatory networks in the latter stage than in the former two stages (Fig. 2C).
A gene ontology (GO) term enrichment analysis was used to elucidate the biological pathways of these DEGs. In stg1, enriched processes included TF activity, sequence-specific DNA binding, nucleic acid binding TF activity, oxylipin biosynthetic and metabolic process and oxidoreductase activity, and involved 65, 65, 7, 7 and 82 genes, respectively (Fig. 2D). However, in stg2, only two categories of TF activity, sequence-specific DNA binding and nucleic acid binding TF activity were significantly enriched, involving 38 and 38 genes, respectively (Fig. 2E). In stg3, the enriched terms were mainly associated with photosynthesis and response to abiotic stimulus (Fig. 2F). Thus, the mechanisms in stg1 and stg2 that underlie CSSL1283 ear-length development were similar but different from those in stg3.
Activated expression of AP2/EREBP and WRKY family genes in CSSL1283
In plants, TFs are important regulators of growth and development. The GO analysis showed that the term of TF activity and sequence-specific DNA binding (GO: 0003700) was significantly enriched in both stg1 and stg2, suggesting that TFs play key roles in CSSL1283 ear-length development at these two stages (Fig. 2D, E). There were 65 and 38 differentially expressed TFs (DE TFs) in stg1 and stg2, respectively (Table S2).
The DE TFs in stg1 included members of the AP2/EREBP, WRKY, bHLH, MADS, HSF, bZIP, C2H2, AHL, GRAS, Homeobox, MYB and GATA families (Table S2). Notably, 41.54% (27/65) and 15.38% (10/65) of the total DE TFs belonged to the AP2/EREBP and WRKY families, respectively (Table S2), revealing that AP2/EREBP and WRKY genes were the major regulators of the increased ear length of CSSL1283. Interestingly, 26 of 27 AP2/EREBP genes were upregulated in CSSL1283 (Fig. 4A). Similarly, 9 of 10 WRKY genes were upregulated in CSSL1283 (Fig. 4C). In stg2, 52.63% (20/38) of the DE TFs belonged to the AP2/EREBP family (Table S2). Consistent with the expression pattern of AP2/EREBP genes in stg1, 19 of 20 AP2/EREBP genes were upregulated in CSSL1283 (Fig. 4B). Thus, the enhanced expression of AP2/EREBP and WRKY TFs appears to be related to the increased ear length of CSSL1283.
Expression profiles of two transcription factor families differentially expressed between Xu178 and CSSL1283 in stg1 and stg2. A and B Heat maps of AP2/EREBP family in stg1 (A) and stg2 (B) ears of Xu178 and CSSL1283. C The heat map of the WRKY family in stg1 ears of Xu178 and CSSL1283. For each gene, FPKM values were normalized using the higher FPKM value of the gene across the two lines. The color scale represents expression levels from high (red) to low (blue color). Stg1-1283 and stg1-178 indicate the stg1 ears of CSSL1283 and Xu178, respectively. Stg2-1283 and stg2-178 indicate the stg2 ears of CSSL1283 and Xu178, respectively
Protein kinase and heat shock protein are required for ear-length development in CSSL1283
A serine/threonine-protein kinase encoding gene, KNR6, was reported to determine pistillate floret number and ear length, suggesting the significance of protein kinases during maize ear-length development (Jia et al. 2020). Thus, DEGs related to protein kinases in stg1 were analyzed in detail. In total, 47 genes belonging to the protein kinase family were differentially expressed between Xu178 and CSSL1283 (Table S3). This included 14 genes encoding receptor-like protein kinases (RLKs) and 7 genes involved in the mitogen-activated protein kinase (MAPK) signaling pathway (Fig. 5A, B). In addition, opposite expression trends for the two kinds of protein kinase genes were observed. In CSSL1283, 12 of 14 DEGs encoding RLKs were downregulated, whereas all the DEGs involved in the MAPK signaling pathway were upregulated (Fig. 5A, B), implying that RLKs and members of the MAPK signaling pathway play different roles in CSSL1283 ear-length development.
Expression profiles of DEGs encoding protein kinases in stg1 and heat shock proteins in stg3. A and B Heat maps of DEGs related to receptor-like protein kinases (A) and the mitogen-activated protein kinase signaling pathway (B) in stg1 ears of Xu178 and CSSL1283. C The heat map of DEGs related to heat shock proteins in stg3 ears of Xu178 and CSSL1283. For each gene, FPKM values were normalized using the higher FPKM value of the gene across the two lines. The color scale represents expression levels from high (red) to low (blue color). Stg1-1283 and stg1-178 indicate the stg1 ears of CSSL1283 and Xu178, respectively. Stg3-1283 and stg3-178 indicate the stg3 ears of CSSL1283 and Xu178, respectively
The GO analysis revealed that the term “response to abiotic stimulus” (GO: 0009628) was the most significantly enriched in stg3 (Fig. 2F). This term included 68 DEGs, with 27 being assigned to the GO term “response to heat” (GO: 0009408) (Fig. 2F). The other DEGs in the term “response to heat”, except for three genes, were upregulated in the CSSL1283 (Table S4). Furthermore, 15 of 27 DEGs encoded heat shock proteins (HSPs), and all of them are upregulated in CSSL1283 (Fig. 5C), suggesting that there is a positive connection between ear-length development and response to heat and that the HSPs are involved in these two biological processes.
Discussion
The TFs play key roles in ear-length development
Several TFs have been reported in maize ear inflorescence development, such as UNBRANED 2, UB3 and FASCIATED EAR4 (Chuck et al. 2014; Pautler et al. 2015). In line with these results, our transcriptomic data revealed that TFs are important for early ear-length development.
AP2/EREBP family genes are involved in the development of IMs in maize. For instance, the SMs fate is controlled by two AP2/EREBP family genes, Indeterminate Spikelet1 and Branched Silkless1 (Chuck et al. 1998; Chuck et al. 2002). Moreover, the increased expression of the former enhances spikelet pair meristem and kernel row numbers (Wang et al. 2019). In the present study, 27 and 20 DEGs of the AP2/EREBP family were detected in stg1 and stg2 (Table S2), respectively. Surprisingly, most of these genes were upregulated in CSSL1283 compared with Xu178 (Fig. 4A, B), indicating that the activated expression of AP2/EREBP family genes may be responsible for the increased ear length of CSSL1283.
Potential roles of protein kinases in ear-length development
Protein phosphorylation by protein kinases can regulate plant growth and environmental responses. In Arabidopsis, most of the protein kinases are RLKs, accounting for approximately 60% of the total protein kinases (Shiu and Bleecker, 2001). In maize, a leucine-rich repeat receptor-like kinase, thick tassel dwarf1, is involved in male and female inflorescence development (Bommert et al. 2005). In addition, the two kernel row number genes, FEA2 and FEA3, encode leucine-rich repeat receptor-like proteins (Bommert et al. 2013b; Je et al. 2016; Taguchi-Shiobara et al. 2001). Because it lacks a signaling domain, the function of FEA2 depends on COMPACT PLANT2, which encodes the predicted α-subunit of a heterotrimeric GTP-binding protein (Bommert et al. 2013a). These findings indicate that signal transduction is essential for ear inflorescence development. In this study, 14 RLKs were differentially expressed in stg1 ear inflorescences between Xu178 and CSSL1283 (Fig. 5A). Thus, we speculate that the signal transduction mediated by RLKs is an important mechanism underling the increased ear length of CSSL1283.
Auxin is a central regulator in ear inflorescence development (Gallavotti et al. 2008; Galli et al. 2015; McSteen et al. 2007; Phillips et al. 2011). Auxin transport can be affected by protein kinases, such as BARREN INFLORESCENCE2 in maize (Skirpan, 2009), and PINOID and MAP KINASE KINASE7 in Arabidopsis (Dai et al. 2006; Friml et al. 2004). During stg1, 7 genes implicated in the MAPK signaling pathway and 8 auxin-related genes were identified as being differentially expressed between Xu178 and CSSL1283 (Fig. 5B, Table S5). Considering the potential function of protein kinase KNR6 in auxin-dependent ear-length development (Jia et al. 2020), it is likely that the cascade of “MAPK signaling pathway-auxin” is a contributor to the increased ear length of CSSL1283.
The ear-length development involves complicated biological processes
Ear length is a typical quantitative trait and is controlled by many genetic loci. Consistently, the GO analysis showed that TFs are the major regulators of ear-length development in stg1 and stg2 (Fig. 2D, E), whereas processes related to photosynthesis and responses to abiotic stimulus played important roles in stg3 (Fig. 2F). This suggested that various biological processes are required for ear-length development in maize.
Adverse environmental conditions, such as high temperature, influence ear development and grain yield. Emerging evidence indicates that HSPs are involved in thermotolerance (Maestri et al. 2002). In addition, HSPs regulate the formation of chalky grains under heat-stress conditions (Tsutomu et al. 2018). HSP90 and a gene encoding a protein kinase WAK may bind to Ca2 + signaling related genes to regulate lipid and nitrogen metabolism, thereby improving salt tolerance in barley (Zhu et al. 2020). WRKY TFs modulate many developmental processes and stress resistance (Rushton et al. 2010; Wang et al. 2018). Thus, HSPs and WRKY TFs have dual functions in plant growth. In our data, the expression levels of DEGs related to HSPs and WRKY TFs were induced in CSSL1283 (Fig. 4C, 5C), suggesting that they can coordinate ear-length development and heat tolerance in maize. Therefore, the identified genes provide key targets for the improvement of maize grain yield.
In addition, EAD1 expression was downregulated in Xu178 in stg1. Overexpression of EAD1 resulted in longer ear length and an increase in kernel number per row (Pei et al. 2022), which is consistent with our results. These results further suggest that the transport and distribution of malate regulates ear development in maize.
Materials and methods
Plant materials and phenotypic analysis
CSSLs were constructed using two maize elite inbred lines, Zong3 and Xu178, which are the donor and recipient parents, respectively (Mao et al. 2013). These lines were planted in the summer of 2019 in the experimental field of Henan Agricultural University in Zhengzhou, Henan Province. At maturity, randomly selected ears were used to measure the ear length and kernel number per row. The observation of young ears was conducted using a Zeiss dissecting microscope. The lengths of young ears were calculated using ZEN 2011 software. The stg1 to stg3 young ears were collected for RNA-seq. This included two biological replicates in stg1 and three biological replicates in stg2 and stg3. The young ears were stored at -80 ℃ for further use.
Total RNA extraction and transcriptome sequencing
The total RNA was extracted from ear inflorescence using an RNAprep Pure Plant Plus Kit (Tiangen). mRNAs were enriched with oligo (dT) magnetic beads and then fragmented into short fragments by adding a fragmentation buffer. The first cDNA strand was synthesized using random hexamers and the associated buffer. Then, dNTPs, RNase H and DNA polymerase I were added to synthesize the second cDNA strand. Double-stranded cDNA was terminal repaired and added to the 3′ ends. The QiaQuick PCR kit was used for purification, and samples were eluted in EB buffer. The ends were repaired and the sequencing adapter was connected. Agarose gel electrophoresis was used to select the appropriate fragment size, and finally, PCR amplification was performed to enrich the cDNA. Library quality was determined using the Agilent 2100 system. The library was then sequenced using Illumina Novaseq to generate 150-nucleotide paired-end reads.
Transcriptomic analysis
All the RNA-seq reads were mapped to the B73 maize reference genome ( B73 AGPv4, http://ensembl.gramene.org/Zea_mays/Info/Index) using Hisat2 (Dijkema et al. 2017). Cufflinks v2.2.12 was used for estimating normalized gene expression values. The differential expression analysis was carried out using Cuffdiff v2.2.13. Genes with an expression fold change ≥ 2 and FDR ≤ 0.05 were defined as differentially expressed. AgriGO v2.0 (http://systemsbiology.cau.edu.cn/agriGOv2/index.php) was used for the GO enrichment analysis(Du et al. 2010).
qRT-PCR
In total, 1 µg RNA was reverse transcribed following the procedures of the PrimeScript™, ® Reagent Kit with gDNA Eraser (TaKaRa). qRT-PCR was performed on a CFX96 Real-Time PCR Detection System (Bio-Rad) using TB Green™ Premix Ex Taq™ II (TaKaRa). Three biological replications were included, and each biological replicate contained three technical replications. The maize Actin gene (Zm00001d010159) was used as the internal normalization control. The gene expression level was evaluated using the 2−ΔΔCt method (Livak and Schmittgen, 2001). All the primers were designed using NCBI (https://www.ncbi.nlm.nih.gov/tools/primer-blast) and are listed in Table S6.
Data availability
The datasets generated during the current study are available in the NCBI Sequence Read Archive (SRA) database under Bio project PRJNA862339 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA862339).
References
Begcy K, Nosenko T, Zhou LZ et al (2019) Male sterility in maize after transient heat stress during the tetrad stage of pollen development. Plant Physiol 181(2):683–700. https://doi.org/10.1104/pp.19.00707
Bommert P, Je BI, Goldshmidt A et al (2013a) The maize Gα gene COMPACT PLANT2 functions in CLAVATA signalling to control shoot meristem size. Nature 502(7472):555–558. https://doi.org/10.1038/nature12583
Bommert P, Lunde C, Nardmann J et al (2005) Thick tassel dwarf1 encodes a putative maize ortholog of the Arabidopsis CLAVATA1 leucine-rich repeat receptor-like kinase. Development 132(6):1235–1245. https://doi.org/10.1242/dev.01671
Bommert P, Nagasawa NS, Jackson D (2013b) Quantitative variation in maize kernel row number is controlled by the FASCIATED EAR2 locus. Nat Genet 45(3):334–337. https://doi.org/10.1038/ng.2534
Chen Z, Liu Y, Yin Y et al (2019) Expression of AtGA2ox1 enhances drought tolerance in maize. Plant Growth Regul 89:203–215. https://doi.org/10.1007/s10725-019-00526-x
Chuck G, Meeley RB, Hake S (1998) The control of maize spikelet meristem fate by the APETALA2-like gene indeterminate spikelet1. Genes and Development 12(8):1145–1154. https://doi.org/10.1101/gad.12.8.1145
Chuck G, Muszynski M, Kellogg E et al (2002) The control of spikelet meristem identity by the branched silkless1 gene in maize. Science 298(5596):1238–1241. https://doi.org/10.1126/science.1076920
Chuck GS, Brown PJ, Meeley R et al (2014) Maize SBP-box transcription factors unbranched2 and unbranched3 affect yield traits by regulating the rate of lateral primordia initiation. Proc Natl Acad Sci USA 111(52):18775–18780. https://doi.org/10.1073/pnas.1407401112
Dai Y, Wang H, Li B et al (2006) Increased expression of MAP KINASE KINASE7 causes deficiency in polar auxin transport and leads to plant architectural abnormality in Arabidopsis. Plant Cell 18(2):308–320. https://doi.org/10.1105/tpc.105.037846
Dijkema EJ, Leiner T, Grotenhuis HB (2017) Diagnosis, imaging and clinical management of aortic coarctation. Heart 103(15):1148–1155. https://doi.org/10.1136/heartjnl-2017-311173
Du X, Fang T, Liu Y et al (2019) Transcriptome profiling predicts new genes to promote maize callus formation and transformation. Front Plant Sci 10:1633. https://doi.org/10.3389/fpls.2019.01633
Du Z, Xin Z, Yi L et al (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38(suppl2):W64–W70. https://doi.org/10.1093/nar/gkq310
Eveland AL, Goldshmidt A, Pautler M et al (2013) Regulatory modules controlling maize inflorescence architecture. Genome Res 24(3):431–443. https://doi.org/10.1101/gr.166397.113
Friml J, Yang X, Michniewicz M et al (2004) A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306(5697):862–865. https://doi.org/10.1126/science.1100618
Gallavotti A, Barazesh S, Malcomber S et al (2008) Sparse inflorescence1 encodes a monocot-specific YUCCA-like gene required for vegetative and reproductive development in maize. Proc Natl Acad Sci USA 105(39):15196–15201. https://doi.org/10.1073/pnas.0805596105
Galli M, Liu Q, Moss BL et al (2015) Auxin signaling modules regulate maize inflorescence architecture. Proc Natl Acad Sci USA 112(43):13372–13377. https://doi.org/10.1073/pnas.1516473112
Godfray HCJ, Beddington JR, Crute IR et al (2010) Food security: the challenge of feeding 9 billion people. Science 327(5967):812–818. https://doi.org/10.1126/science.1185383
Huo D, Ning Q, Shen X et al (2016) QTL mapping of kernel number-related traits and validation of one major QTL for ear length in maize. PLoS ONE 11(5):e0155506. https://doi.org/10.1371/journal.pone.0155506
Je BI, Gruel J, Lee YK et al (2016) Signaling from maize organ primordia via FASCIATED EAR3 regulates stem cell proliferation and yield traits. Nat Genet 48(7):785–791. https://doi.org/10.1038/ng.3567
Jia H, Li M, Li W et al (2020) A serine/threonine protein kinase encoding gene KERNEL NUMBER PER ROW6 regulates maize grain yield. Nat Commun 11(1):1–11. https://doi.org/10.1038/s41467-020-14746-7
Liu L, Du Y, Shen X et al (2015) KRN4 controls quantitative variation in maize kernel row number. PLoS Genet 11(11):e1005670. https://doi.org/10.1371/journal.pgen.1005670
Liu R, Jia H, Cao X et al (2012) Fine mapping and candidate gene prediction of a pleiotropic quantitative trait locus for yield-related trait in Zea mays. PLoS ONE 7(11):e49836. https://doi.org/10.1371/journal.pone.0049836
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR the 2–∆∆Ct method. Methods 25(4):402–408. https://doi.org/10.1006/meth.2001.1262
Luo Y, Zhang M, Liu Y et al (2022) Genetic variation in YIGE1 contributes to ear length and grain yield in maize. New Phytol 234(2):513–526. https://doi.org/10.1111/nph.17882
Ma X, Tang J, Teng W et al (2007) Epistatic interaction is an important genetic basis of grain yield and its components in maize. Mol Breeding 20(1):41–51. https://doi.org/10.1007/s11032-006-9071-9
Maestri E, Klueva N, Perrotta C et al (2002) Molecular genetics of heat tolerance and heat shock proteins in cereals. Plant Mol Biol 48(5–6):667–681. https://doi.org/10.1023/A:1014826730024
Mao K, Li W, Fu Z et al (2013) Development of a set of single segment substitution lines of an elite inbred line Zong 3 on the genetic background Xu 178 in maize (Zea mays L.). J Henan Agricultural Univ 47(1):6–915. https://doi.org/10.3969/j.issn.1000-2340.2013.01.002
McSteen P, Malcomber S, Skirpan A et al (2007) Barren inflorescence2 encodes a co-ortholog of the PINOID serine/threonine kinase and is required for organogenesis during inflorescence and vegetative development in maize. Plant Physiol 144(2):1000–1011. https://doi.org/10.1104/pp.107.098558
Ning Q, Jian Y, Du Y et al (2021) An ethylene biosynthesis enzyme controls quantitative variation in maize ear length and kernel yield. Nat Commun 12(1):1–10. https://doi.org/10.1038/s41467-021-26123-z
Pautler M, Eveland AL, Larue T et al (2015) FASCIATED EAR4 encodes a bZIP transcription factor that regulates shoot meristem size in maize. Plant Cell 27(1):104–120. https://doi.org/10.1105/tpc.114.132506
Pei Y, Deng Y, Zhang H et al (2022) EAR APICAL DEGENERATION1 regulates maize ear development by maintaining malate supply for apical inflorescence. Plant Cell 62222–2241. https://doi.org/10.1093/plcell/koac093
Phillips KA, Skirpan AL, Liu X et al (2011) vanishing tassel2 encodes a grass-specific tryptophan aminotransferase required for vegetative and reproductive development in maize. The Plant Cell 23(2):550–566. https://doi.org/10.1105/tpc.110.075267
Rushton PJ, Somssich IE, Ringler P et al (2010) WRKY transcription factors. Trends Plant Sci 15(5):247–258. https://doi.org/10.4161/psb.27700
Shiu SH, Bleecker AB (2001) Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci USA 98(19):10763–10768. https://doi.org/10.1073/pnas.181141598
Skirpan A, Culler A, Gallavotti A et al (2009) BARREN INFLORESCENCE2 interaction with ZmPIN1a suggests a role in auxin transport during maize inflorescence development. Plant Cell Physiol 50(3):652–657. https://doi.org/10.1093/pcp/pcp006
Taguchi-Shiobara F, Yuan Z, Hake S et al (2001) The fasciated ear2 gene encodes a leucine-rich repeat receptor-like protein that regulates shoot meristem proliferation in maize. Genes and Development 15(20):2755–2766. https://doi.org/10.1101/gad.208501
Vollbrecht E, Schmidt RJ (2009) Development of the inflorescences. In: Handbook of maize: Its biology 13–40. https://doi.org/10.1007/978-0-387-79418-1_2
Wang C, Ru J, Liu Y et al (2018) Maize WRKY transcription factor ZmWRKY106 confers drought and heat tolerance in transgenic plants. Int J Mol Sci 19(10):3046. https://doi.org/10.3390/ijms19103046
Wang J, Lin Z, Zhang X et al (2019) krn1, a major quantitative trait locus for kernel row number in maize. New Phytol 223(3):1634–1646. https://doi.org/10.1111/nph.15890
Xiao Y, Tong H, Yang X et al (2016) Genome-wide dissection of the maize ear genetic architecture using multiple populations. New Phytol 210(3):1095–1106. https://doi.org/10.1111/nph.13814
Yang N, Lu Y, Yang X et al (2014) Genome wide association studies using a new nonparametric model reveal the genetic architecture of 17 agronomic traits in an enlarged maize association panel. PLoS Genet 10(9):e1004573. https://doi.org/10.1371/journal.pgen.1004573
Yi F, Gu W, Chen J et al (2019) High temporal-resolution transcriptome landscape of early maize seed development. Plant Cell 31(5):974–992. https://doi.org/10.1105/tpc.18.00961
Zhao J, He Y, Li X et al (2020) An integrated RNA-Seq and physiological study reveals gene responses involving in the initial imbibition of seed germination in rice. Plant Growth Regul 90:249–263. https://doi.org/10.1007/s10725-019-00567-2
Zhou G, Zhu Q, Yang G et al (2015) qEL7.2 is a pleiotropic QTL for kernel number per row, ear length and ear weight in maize (Zea mays L.). Euphytica 203(2):429–436. https://doi.org/10.1007/s10681-014-1307-x
Zhu J, Fan Y, Li C et al (2020) Candidate genes for salinity tolerance in barley revealed by RNA-seq analysis of near-isogenic lines. Plant Growth Regul 92:571–582. https://doi.org/10.1007/s10725-020-00662-9
Acknowledgements
This work was supported by National Natural Science Foundation of China (No. 31801379, 31971961). Thanks Liwen Bianji (Edanz) (https://www.liwenbianji.cn) for editing the language of a draft of this manuscript.
Funding
Funding was provided by National Natural Science Foundation of China (Grant No.: 31801379, 31971961)
Author information
Authors and Affiliations
Contributions
All authors contributed to the study’s conception and design. Xiaoyang Chen, Dong Ding and Jihua Tang designed the research. Shujun Meng, Yujie Lian, Hui Chen and Xudong Cao performed the experiments. Yuming Huang and Shujun Meng analyzed the data. Xiaoyang Chen and Shujun Meng wrote the manuscript, Dong Ding and Jihua Tang revised the manuscript. All authors reviewed the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
All experimental studies on plants have complied with relevant institutional, national, and international guidelines and legislation.
Additional information
Communicated by Ben Zhang.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Meng, S., Huang, Y., Lian, Y. et al. Transcriptomic analysis reveals the regulation of early ear-length development in maize. Plant Growth Regul 100, 97–105 (2023). https://doi.org/10.1007/s10725-022-00941-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-022-00941-7