Abstract
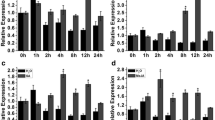
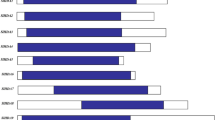
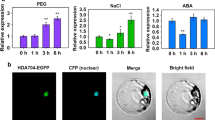
Histone deacetylation, one of the vital modifying factors of post-translational modifications, is catalyzed by histone deacetylase. The genes of histone deacetylase (HDACs) play critical roles in responses to various stress. However, the detailed functions of most SlHDAC members in tomato remain unknown. In this work, we found that a histone deacetylase, SlHDA3, is involved in the response to NaCl and drought abiotic stresses. The expression of SlHDA3 was also induced significantly by NaCl, drought stress and hormone treatments. Upon silencing of SlHDA3 in tomato, the RNAi transgenic plants presented depressed tolerance to drought and salt stresses compared with Wild-type (WT) tomato. The results of the sensitivity experiment analysis indicated that the length of hypocotyl and root in RNAi plants were more inhibited by ABA and salt stress compared to those in WT plants at the post-germination stage. More serious growth status was exhibited in SlHDA3 transgenic plants under salt and drought stress, as evaluated by a series of physiological parameters related to stress responses, such as decreased Survival ratio (RWC), survival rate, Abscisic acid (ABA) content, chlorophyll content and Catalase (CAT) activity, and increased Malondialdehyde (MDA) and proline contents. In addition, the expression analysis of transgenic plants by quantitative reverse transcription-PCR (qRT-PCR) showed that the transcripts of genes associated with responses to abiotic stress were down-regulated under salt-stressed conditions. In summary, SlHDA3 acts as a stress-responsive gene, plays a role in the positive regulation of abiotic stress tolerance, and may be one of the new target genes in the engineering breeding of salt- and drought-tolerant tomato.








Similar content being viewed by others
Abbreviations
- HDACs:
-
Histone deacetylases
- HATs:
-
Histone acetyltransferases
- ABA:
-
Abscisic acid
- CAT:
-
Catalase
- GA:
-
Gibberellin acid
- IAA:
-
Indole-3-acetic acid
- MDA:
-
Malondialdehyde
- Pro:
-
Proline
- qRT-PCR:
-
quantitative reverse transcription-PCR
- ROS:
-
Reactive oxygen species
- SA:
-
Salicylic acid
- WT:
-
Wild-type
- RWC:
-
Relative water content
References
Abdelkareem A, Thagun C, Nakayasu M, Mizutani M, Hashimoto T, Shoji T (2017) Jasmonate-induced biosynthesis of steroidal glycoalkaloids depends on COI1 proteins in tomato. Biochem Biophys Res Commun 489(2):206–210. https://doi.org/10.1016/j.bbrc.2017.05.132
Aoki K, Yano K, Suzuki A, Kawamura S, Sakurai N, Suda K, Kurabayashi A, Suzuki T, Tsugane T, Watanabe M, Ooga K, Torii M, Narita T, Shin IT, Kohara Y, Yamamoto N, Takahashi H, Watanabe Y, Egusa M, Kodama M, Ichinose Y, Kikuchi M, Fukushima S, Okabe A, Arie T, Sato Y, Yazawa K, Satoh S, Omura T, Ezura H, Shibata D (2010) Large-scale analysis of full-length cDNAs from the tomato (Solanum lycopersicum) cultivar Micro-Tom, a reference system for the Solanaceae genomics. BMC Genomics 11:210. https://doi.org/10.1186/1471-2164-11-210
Aufsatz W, Mette MF, van der Winden J, Matzke M, Matzke AJM (2002) HDA6, a putative histone deacetylase needed to enhance DNA methylation induced by double-stranded RNA. EMBO 21(24):6832–6841. https://doi.org/10.1093/emboj/cdf663
Benhamed M, Bertrand C, Servet C, Zhou DX (2006) Arabidopsis GCN5, HD1, and TAF1/HAF2 interact to regulate histone acetylation required for light-responsive gene expression. Plant Cell 18(11):2893–2903
Bolger A, Scossa F, Bolger ME, Lanz C, Maumus F, Tohge T, Quesneville H, Alseekh S, Sørensen I, Lichtenstein G, Fich EA, Conte M, Keller H, Schneeberger K, Schwacke R, Ofner I, Vrebalov J, Xu Y, Osorio S, Aflitos SA, Schijlen E, Jiménez-Goméz JM, Ryngajllo M, Kimura S, Kumar R, Koenig D, Headland LR, Maloof JN, Sinha N, van Ham RCHJ, Lankhorst RK, Mao L, Vogel A, Arsova B, Panstruga R, Fei Z, Rose JKC, Zamir D, Carrari F, Giovannoni JJ, Weigel D, Usadel B, Fernie AR (2014) The genome of the stress-tolerant wild tomato species Solanum pennellii. Nat Genet 46(9):1034–1038. https://doi.org/10.1038/ng.3046
Buszewicz D, Archacki R, Palusiński A, Kotliński M, Fogtman A, Iwanicka-Nowicka R, Sosnowska K, Kuciński J, Pupel P, Olędzki J, Dadlez M, Misicka A, Jerzmanowski A, Koblowska MK (2016) HD2C histone deacetylase and a SWI/SNF chromatin remodelling complex interact and both are involved in mediating the heat stress response in Arabidopsis. Plant Cell Environ 39(10):2108–2122. https://doi.org/10.1111/pce.12756
Cardenas PD, Sonawane PD, Pollier J, Vanden Bossche R, Dewangan V, Weithorn E, Tal L, Meir S, Rogachev I, Malitsky S, Giri AP, Goossens A, Burdman S, Aharoni A (2016) GAME9 regulates the biosynthesis of steroidal alkaloids and upstream isoprenoids in the plant mevalonate pathway. Nat Commun 7:10654. https://doi.org/10.1038/ncomms10654
Chen LT, Wu K (2010) Role of histone deacetylases HDA6 and HDA19 in ABA and abiotic stress response. Plant Signal Behav 5(10):1318–1320
Chen LT, Luo M, Wang YY, Keqiang W (2010) Involvement of Arabidopsis histone deacetylase HDA6 in ABA and salt stress response. J Exp Bot 61(12):3345–3353
Chen P, Sun YF, Kai WB, Liang B, Zhang YS, Zhai XW, Jiang L, Du YW, Leng P (2016) Interactions of ABA signaling core components (SlPYLs, SlPP2Cs, and SlSnRK2s) in tomato (Solanum lycopersicon). J Plant Physiol. https://doi.org/10.1016/j.jplph.2016.07.016
Chinnusamy V, Gong Z, Zhu JA (2008) Abscisic acid-mediated epigenetic processes in plant development and stress responses. J Integr Plant Biol 50(10):1187–95
Danquah A, De Zelicourt A, Colcombet J, Hirt H (2014) The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnol Adv 32(1):40–52
Fujita M, Fujita Y, Maruyama K, Seki M, Hiratsu K, Ohme-Takagi M, Tran LSP, Yamaguchi-Shinozaki K, Shinozaki K (2010) A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J 39(6):863–876
Gaffe J, Lemercier C, Alcaraz JP, Kuntz M (2011) Identification of three tomato flower and fruit MADS-box proteins with a putative histone deacetylase binding domain. Gene 471:19–26
Georgieva EI, López-Rodas G, Sendra R, Grbner P, Loidl P (1991) Histone acetylation in Zea mays II: biological significance of post-translational histone acetylation during embryo germination. J Biol Chem 266(28):18751–18760
Gonzalez D, Bowen AJ, Carroll TS, Conlan RS (2007) The transcription corepressor leunig interacts with the histone deacetylase HDA19 and mediator components MED14 (SWP) and CDK8 (HEN3) to repress transcription. Mol Cell Biol 27(15):5306–5315
Gu X, Jiang D, Yang W, Jacob Y, Michaels SD, He Y, Chen X (2011) Arabidopsis homologs of retinoblastoma-associated protein 46/48 associate with a histone deacetylase to act redundantly in chromatin silencing. PLoS Genet 7(11):e1002366
Guo JE (2022) Histone deacetylase gene SlHDT1 regulates tomato fruit ripening by affecting carotenoid accumulation and ethylene biosynthesis. Plant Sci 318:111235. https://doi.org/10.1016/j.plantsci.2022.111235
Guo JE, Hu Z, Li F, Zhang L, Yu X, Tang B, Chen G (2017a) Silencing of histone deacetylase SlHDT3 delays fruit ripening and suppresses carotenoid accumulation in tomato. Plant Sci 265:29–38
Guo JE, Hu Z, Guo X, Zhang L, Yu X, Zhou S, Chen G (2017b) Molecular characterization of nine tissue-specific or stress-responsive genes of histone deacetylase in tomato (Solanum lycopersicum). J Plant Growth Regul. 36(3):566–577
Guo JE, Hu Z, Zhu M, Li F, Zhu Z, Lu Y, Chen G (2017c) The tomato histone deacetylase SlHDA1 contributes to the repression of fruit ripening and carotenoid accumulation. Sci Rep 7(1):7930. https://doi.org/10.1038/s41598-017-08512-x
Guo JE, Hu Z, Yu X, Li A, Li F, Wang Y, Tian S, Chen G (2018) A histone deacetylase gene, SlHDA3, acts as a negative regulator of fruit ripening and carotenoid accumulation. Plant Cell Rep 37(1):125–135
Itkin M, Heinig U, Tzfadia O, Bhide AJ, Shinde B, Cardenas PD, Bocobza SE, Unger T, Malitsky S, Finkers R, Tikunov Y, Bovy A, Chikate Y, Singh P, Rogachev I, Beekwilder J, Giri AP, Aharoni A (2013) Biosynthesis of antinutritional alkaloids in solanaceous crops is mediated by clustered genes. Sci 341(6142):175–179. https://doi.org/10.1126/science.1240230
Ji K, Kai W, Zhao B, Sun Y, Yuan B, Dai S, Li Q, Chen P, Wang Y, Pei Y (2014) SlNCED1 and SlCYP707A2: key genes involved in ABA metabolism during tomato fruit ripening. J Exp Bot 18:5243–5255
Jong-Myong K, Kim TT, Junko I, Akihiro M, Hiroshi K, Motoaki S (2012) Transition of chromatin status during the process of recovery from drought stress in Arabidopsis thaliana. Plant Cell Physiol 53(5):847
Kim JM, To TK, Ishida J, Morosawa T, Kawashima M, Matsui A (2008) Alterations of lysine modifications on the histone H3 N-tail under drought stress conditions in Arabidopsis thaliana. Plant Cell Physiol. https://doi.org/10.1093/pcp/pcn133
Kim W, Latrasse D, Servet C, Zhou DX (2013) Arabidopsis histone deacetylase HDA9 regulates flowering time through repression of AGL19. Biochem Biophys Res Commun 432(2):394–398
Kumar V, Thakur JK, Prasad M (2021) Histone acetylation dynamics regulating plant development and stress responses. Cell Mol Life Sci 78(10):4467–4486
Lei B, Berger F (2020) H2A variants in Arabidopsis: versatile regulators of genome activity. Plant Commun 1(1):100015
Li J, Chitwood J, Menda N, Mueller L, Hutton SF (2018) Linkage between the I-3 gene for resistance to Fusarium wilt race 3 and increased sensitivity to bacterial spot in tomato. Theor Appl Genet 131(1):145–155. https://doi.org/10.1007/s00122-017-2991-4
Liu C, Li LC, Chen WQ, Chen X, Xu ZH, Bai SN (2013) HDA18 affects cell fate in Arabidopsis root epidermis via histone acetylation at four kinase genes. Plant Cell 25(1):257–269. https://doi.org/10.1105/tpc.112.107045
Liu XC, Yang SG, Zhao ML, Luo CW, Chen CY (2014) Transcriptional repression by histone deacetylases in plants. Mol Plant. 7(5):764–772
Luo M, Liu X, Singh P, Cui Y, Zimmerli L, Wu K (2012a) Chromatin modifications and remodeling in plant abiotic stress responses. Biochim Biophys Acta 1819(2):129–136
Luo M, Wang YY, Liu X, Yang S, Wu K (2012b) HD2C interacts with HDA6 and is involved in ABA and salt stress response in Arabidopsis. J Exp Bot 63(8):3297–3306
Luo M, Yu CW, Chen FF, Zhao L, Tian G, Liu X, Cui Y, Yang JY, Wu K (2012c) Histone deacetylase HDA6 is functionally associated with AS1 in repression of KNOX genes in Arabidopsis. PLoS Genet 8(12):e1003114. https://doi.org/10.1371/journal.pgen.1003114
Mathews H, Clendennen SK, Caldwell CG, Liu XL, Connors K, Matheis N, Schuster DK, Menasco DJ, Wagoner W, Lightner J, Wagner DR (2003) Activation tagging in tomato identifies a transcriptional regulator of anthocyanin biosynthesis, modification, and transport. Plant Cell 15(8):1689–1703. https://doi.org/10.1105/tpc.012963
Matsuba Y, Nguyen TT, Wiegert K, Falara V, Gonzales-Vigil E, Leong B, Schafer P, Kudrna D, Wing RA, Bolger AM, Usadel B, Tissier A, Fernie AR, Barry CS, Pichersky E (2013) Evolution of a complex locus for terpene biosynthesis in solanum. Plant Cell 25(6):2022–2036. https://doi.org/10.1105/tpc.113.111013
Ming L (2015) Regulation of flowering time by the histone deacetylase HDA5 in Arabidopsis. Plant J 82(82):925–936
Nakashima K, Takasaki H, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2012) NAC transcription factors in plant abiotic stress responses. BBA - Gene Regulatory Mechanisms 1819(2):97–103
Nakayasu M, Shioya N, Shikata M, Thagun C, Abdelkareem A, Okabe Y, Ariizumi T, Arimura GI, Mizutani M, Ezura H, Hashimoto T, Shoji T (2018) JRE4 is a master transcriptional regulator of defense-related steroidal glycoalkaloids in tomato. Plant J 94(6):975–990. https://doi.org/10.1111/tpj.13911
Nicot N, Hausman JF, Hoffmann L, Evers D (2005) Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J Exp Bot 56(421):2907–2914. https://doi.org/10.1093/jxb/eri285
Nitsch LM, Oplaat C, Feron R, Ma Q, Wolters-Arts M, Hedden P, Mariani C, Vriezen WH (2009) Abscisic acid levels in tomato ovaries are regulated by LeNCED1 and SlCYP707A1. Planta 229(6):1335–1346. https://doi.org/10.1007/s00425-009-0913-7
Ofori PA, Mizuno A, Suzuki M, Martinoia E, Reuscher S, Aoki K, Shibata D, Otagaki S, Matsumoto S, Shiratake K (2018) Genome-wide analysis of ATP binding cassette (ABC) transporters in tomato. PLoS ONE 13(7):e0200854. https://doi.org/10.1371/journal.pone.0200854
Ohnishi T, Nomura T, Watanabe B, Ohta D, Yokota T, Miyagawa H, Sakata K, Mizutani M (2006) Tomato cytochrome P450 CYP734A7 functions in brassinosteroid catabolism. Phytochemistry 67(17):1895–1906. https://doi.org/10.1016/j.phytochem.2006.05.042
Perrella G, Lopez-Vernaza MA, Carr C, Sani E (2013) Histone deacetylase complex1 expression level titrates plant growth and abscisic acid sensitivity in Arabidopsis. Plant Cell 25(9):3491–3505
Pri-Hadash A, Hareven D, Lifschitz E (1992) A meristem-related gene from tomato encodes a dUTPase: analysis of expression in vegetative and floral meristems. Plant Cell 4(2):149–159
Scofield S, Murray JAH (2006) KNOX gene function in plant stem cell niches. Plant Mol Biol 60(6):929–946
Song CP (2005) Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell 17(8):2384–2396
Sridha S, Wu K (2006) Identification of AtHD2C as a novel regulator of abscisic acid responses in Arabidopsis. Plant J. 46(1):124–33
Tahir MS, Tian L (2021) HD2-type histone deacetylases: unique regulators of plant development and stress responses. Plant Cell Rep 40:1603–1615
Tanaka M, Kikuchi A, Kamada H (2008) The Arabidopsis histone deacetylases HDA6 and HDA19 contribute to the repression of embryonic properties after germination. Plant Physiol 146(1):149–161
Tang D, Gallusci P, Lang Z (2020) Fruit development and epigenetic modifications. New Phytol 228(3):839–844
Tessadori F, van Zanten M, Pavlova P, Clifton R, Pontvianne F, Snoek LB, Millenaar FF, Schulkes RK, van Driel R, Voesenek LACJ, Spillane C, Pikaard CS, Fransz P, Peeters AJM (2009) Phytochrome B and Histone deacetylase 6 control light-induced chromatin compaction in Arabidopsis thaliana. PLoS Genet 5(9):e1000638. https://doi.org/10.1371/journal.pgen.1000638
Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J (2007) JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature 448(7154):661–665. https://doi.org/10.1038/nature05960
Tian L, Wang J, Fong MP, Chen M, Cao H, Gelvin SB, Chen ZJ (2003) Genetic control of developmental changes induced by disruption of Arabidopsis histone deacetylase 1 (AtHD1) expression. Genetics 165(1):399
Tian L, Song T, He R, Zeng Y, Xie W, Wu Q, Wang S, Zhou X, Zhang Y (2017) Genome-wide analysis of ATP-binding cassette (ABC) transporters in the sweetpotato whitefly, Bemisia tabaci. BMC Genomics 18(1):330. https://doi.org/10.1186/s12864-017-3706-6
To TK, Kim JM (2013) Epigenetic regulation of gene responsiveness in Arabidopsis. Front Plant Sci. https://doi.org/10.3389/fpls.2013.00548
To TK, Kim JM, Matsui A, Kurihara Y, Morosawa T, Ishida J, Tanaka M, Endo T, Kakutani T, Toyoda T (2011) Arabidopsis HDA6 regulates locus-directed heterochromatin silencing in cooperation with MET1. PLoS Genet 7(4):e1002055
Tran LSP, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Yamaguchi-Shinozaki SK (2004) Isolation and functional analysis of arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16(9):2481–2498
Varotto S, Locatelli S, Canova S, Pipal A, Motto M, Rossi V (2003) Expression profile and cellular localization of maize Rpd3-type histone deacetylases during plant development. Plant Physiol 133(2):606–617
Vasav AP, Barvkar VT (2019) Phylogenomic analysis of cytochrome P450 multigene family and their differential expression analysis in Solanum lycopersicum L. suggested tissue specific promoters. BMC Genomics 20(1):116. https://doi.org/10.1186/s12864-019-5483-x
Wang L, Kim J, Somers DE (2012) Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proceedings of the National Academy of Sciences
Wang Z, Cao H, Chen F, Liu Y (2014) The roles of histone acetylation in seed performance and plant development. Plant Physiol Biochem 84:125–133. https://doi.org/10.1016/j.plaphy.2014.09.010
Wu K, Zhang L, Zhou C, Yu CW (2008) HDA6 is required for jasmonate response, senescence and flowering in Arabidopsis. J Exp Bot 59(2):225–234
Yang X, Zhang X, Yang Y, Zhang H, Zhu W, Nie WF (2021) The histone variant Sl_H2A. Z regulates carotenoid biosynthesis and gene expression during tomato fruit ripening. Hortic Res 8(1):85
Yu CW, Liu X, Luo M, Chen C, Lin X (2011) Histone deacetylase6 Interacts with flowering locus D and regulates flowering in Arabidopsis. Plant Physiol. https://doi.org/10.1104/pp.111.174417
Yuan L, Liu X, Luo M, Yang S, Wu K (2013) Involvement of histone modifications in plant abiotic stress responses. J Integr Plant Biol 55(10):892–901
Zhao L, Lu J, Zhang J, Wu PY, Yang S, Wu K (2014a) Identification and characterization of histone deacetylases in tomato (Solanum lycopersicum). Front Plant Sci 5:760
Zhao J, Zhang J, Zhang W, Wu K, Zheng F, Tian L, Liu X, Duan J (2014b) Expression and functional analysis of the plant-specific histone deacetylase HDT701 in rice. Front Plant Sci 5:764. https://doi.org/10.3389/fpls.2014.00764
Zhaofen H, Huimin Y, Zhong Z, David H, Xinjuan L, Jun D, Lining T (2016) AtHD2D gene plays a role in plant growth, development, and response to abiotic stresses in Arabidopsis thaliana. Front Plant Sci 7:114
Zhou J, Wang X, Jiao Y, Qin Y, Liu X, He K, Chen C, Ma L, Wang J, Xiong L, Zhang Q, Fan L, Deng XW (2007) Global genome expression analysis of rice in response to drought and high-salinity stresses in shoot, flag leaf, and panicle. Plant Mol Biol 63(5):591–608
Zhou L, Tian S, Qin G (2019) RNA methylomes reveal the m6A-mediated regulation of DNA demethylase gene SlDML2 in tomato fruit ripening. Genome Biol 20(1):1–23
Zhu Z, An F, Feng Y, Li P, Xue L, Mu A, Jiang Z, Kim JM, To TK, Li W (2011) Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc Natl Acad Sci U S A 108(30):12539–12544
Zhu M, Meng X, Cai J, Li G, Dong T, Li Z (2018) Basic leucine zipper transcription factor SlbZIP1 mediates salt and drought stress tolerance in tomato. BMC Plant Biol 18(1):83. https://doi.org/10.1186/s12870-018-1299-0
Funding
This work is supported by the Shanxi Province Basic Research Plan (No. 202103021224319 and No. 202103021223384), the Natural Science Foundation of Jiangxi Province (No. 20202BABL215009), and 2019 Lvliang Development Zone’s plan to introduce high-level scientific and technological talents(No. 2019101 and No. 2019106).
Author information
Authors and Affiliations
Contributions
JG: conducted experiments, wrote the manuscript and revised the manuscript. HW: conceived and designed research. JL and YY: analyzed data. ZZ: contributed new reagents and plant materials. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Guo, JE., Wang, H., Yang, Y. et al. Histone deacetylase gene SlHDA3 is involved in drought and salt response in tomato. Plant Growth Regul 99, 359–372 (2023). https://doi.org/10.1007/s10725-022-00913-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-022-00913-x