Abstract
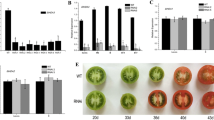
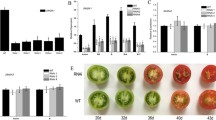
Histone acetylation and deacetylation play an important role in plant growth and development by chromatin modifications. Regulation of histone acetylation and deacetylation is controlled by histone acetyltransferases and histone deacetylases (HDACs) in different tissues and development stages. Knowledge of the importance of genome stability, transcriptional regulation, development and response to stress has been obtained from the model plant Arabidopsis. However, little information on biological functions and development or stress-related HDAC genes is available in tomato. In this study, nine tomato histone deacetylase genes of the RPD3/HDA1 subfamily, from SlHDA1-SlHDA9, were characterized to encode histone deacetylase proteins that share high similarity of protein sequences and conserved domains with those of known plant HDACs. These HDAC genes have been further subdivided into four groups, namely Class I, Class II, Class III and Class IV based on phylogenetic analysis. Quantitative RT-PCR analysis revealed that the nine SlHDAC genes were expressed in all tissues with different transcript abundance and exhibited different tissue-specific expression profiles, suggesting that they may have crucial and diverse roles in tomato growth and development. Varying degrees of induction were detected in the transcript levels of these SlHDACs under various abiotic stresses including NaCl, dehydration, and high/low temperature. These nine SlHDACs were induced by the above stresses with differential/similar induction levels. Overall, this study provides valuable information for further exploring the regulation of HDAC genes during tomato development and fruit ripening and in response to environmental stresses.







Similar content being viewed by others
References
Aiese Cigliano R, Sanseverino W, Cremona G, Ercolano MR, Conicella C, Consiglio FM (2013) Genome-wide analysis of histone modifiers in tomato: gaining an insight into their developmental roles. BMC Genom 14:57–57
Aufsatz W, Mette MF, van der Winden J, Matzke M, Matzke AJM (2002) HDA6, a putative histone deacetylase needed to enhance DNA methylation induced by double-stranded RNA. Embo J 21:6832–6841
Campos EI, Reinberg D (2009) Histones: annotating chromatin. Annu Rev Genet 43:559–599
Chung PJ, Kim YS, Jeong JS, Park S-H, Nahm BH, Kim J-K (2009) The histone deacetylase OsHDAC1 epigenetically regulates the OsNAC6 gene that controls seedling root growth in rice. Plant J 59:764–776
Cigliano RA, Cremona G, Paparo R, Termolino P, Perrella G, Gutzat R, Consiglio MF, Conicella C (2013a) Histone deacetylase AtHDA7 Is required for female gametophyte and embryo development in Arabidopsis. Plant Physiol 163:431–440
Cigliano RA, Sanseverino W, Cremona G, Ercolano MR, Conicella C, Consiglio FM (2013b) Genome-wide analysis of histone modifiers in tomato: gaining an insight into their developmental roles. BMC Genom 14
Costa F, Alba R, Schouten H, Soglio V, Gianfranceschi L, Serra S, Musacchi S, Sansavini S, Costa G, Fei Z, Giovannoni J (2010) Use of homologous and heterologous gene expression profiling tools to characterize transcription dynamics during apple fruit maturation and ripening. BMC Plant Biol 10:1
Durrin LK, Mann RK, Kayne PS, Grunstein M (1991) Yeast histone H4 N-terminal sequence is required for promoter activation in vivo. Cell 65:1023–1031
Exposito-Rodriguez M, Borges AA, Borges-Perez A, Perez JA (2008) Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol 8:1
Frye RA (2000) Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun 273:793–798
Gao MJ, Li X, Huang J, Gropp GM, Gjetvaj B, Lindsay DL, Wei S, Coutu C, Chen Z, Wan XC, Hannoufa A, Lydiate DJ, Gruber MY, Chen ZJ, Hegedus DD (2015) SCARECROW-LIKE15 interacts with HISTONE DEACETYLASE19 and is essential for repressing the seed maturation programme. Nat Commun 6:7243
Gonzalez D, Bowen AJ, Carroll TS, Conlan RS (2007) The transcription corepressor LEUNIG interacts with the histone deacetylase HDA19 and mediator components MED14 (SWP) and CDK8 (HEN3) to repress transcription. Mol Cell Biol 27:5306–5315
Hollender C, Liu Z (2008) Histone deacetylase genes in Arabidopsis development. J Integr Plant Biol 50:875–885
Hristova E, Fal K, Klemme L, Windels D, Bucher E (2015) HISTONE DEACETYLASE6 controls gene expression patterning and DNA methylation-independent euchromatic silencing. Plant Physiol 168:1298–1308
Hu Y, Qin F, Huang L, Sun Q, Li C, Zhao Y, Zhou D-X (2009) Rice histone deacetylase genes display specific expression patterns and developmental functions. Biochem Biophys Res Commun 388:266–271
Kang MJ, Jin HS, Noh YS, Noh B (2015) Repression of flowering under a noninductive photoperiod by the HDA9-AGL19-FT module in Arabidopsis. New Phytol 206:281–294
Kim W, Latrasse D, Servet C, Zhou D-X (2013) Arabidopsis histone deacetylase HDA9 regulates flowering time through repression of AGL19. Biochem Biophys Res Commun 432:394–398
Lee K, Park OS, Jung SJ, Seo PJ (2016) Histone deacetylation-mediated cellular dedifferentiation in Arabidopsis. J Plant Physiol 191:95–100
Li DX, Chen WQ, Xu ZH, Bai SN (2015) HISTONE DEACETYLASE6-defective mutants show increased expression and acetylation of ENHANCER OF TRIPTYCHON AND CAPRICE1 and GLABRA2 with Small but significant effects on root epidermis cellular pattern. Plant Physiol 168:1448–1458
Liu X, Luo M, Zhang W, Zhao J, Zhang J, Wu K, Tian L, Duan J (2012) Histone acetyltransferases in rice (Oryza sativa L.): phylogenetic analysis, subcellular localization and expression. BMC Plant Biol 12:1
Liu X, Chen C-Y, Wang K-C, Luo M, Tai R, Yuan L, Zhao M, Yang S, Tian G, Cui Y, Hsieh H-L, Wu K (2013) PHYTOCHROME INTERACTING FACTOR3 associates with the histone deacetylase HDA15 in repression of chlorophyll biosynthesis and photosynthesis in etiolated Arabidopsis seedlings. Plant Cell 25:1258–1273
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(−Delta Delta C) method. Methods 25:402–408
Long JA, Ohno C, Smith ZR, Meyerowitz EM (2006) TOPLESS regulates apical embryonic fate in Arabidopsis. Science 312:1520–1523
Luo M, Liu X, Singh P, Cui Y, Zimmerli L, Wu K (2012a) Chromatin modifications and remodeling in plant abiotic stress responses. Biochim Biophys Acta 1819:129–136
Luo M, Wang Y-Y, Liu X, Yang S, Lu Q, Cui Y, Wu K (2012b) HD2C interacts with HDA6 and is involved in ABA and salt stress response in Arabidopsis. J Exp Bot 63:3297–3306
Lusser A, Brosch G, Loidl A, Haas H, Loidl P (1997) Identification of maize histone deacetylase HD2 as an acidic nucleolar phosphoprotein. Science 277:88–91
Manning K, Tor M, Poole M, Hong Y, Thompson AJ, King GJ, Giovannoni JJ, Seymour GB (2006) A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat Genet 38:948–952
Nakashima K, Ito Y, Yamaguchi-Shinozaki K (2009) Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol 149:88–95
Nicot N, Hausman JF, Hoffmann L, Evers D (2005) Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J Exp Bot 56:2907–2914
Pan Y, Seymour GB, Lu C, Hu Z, Chen X, Chen G (2012) An ethylene response factor (ERF5) promoting adaptation to drought and salt tolerance in tomato. Plant Cell Rep 31:349–360
Pandey R, Muller A, Napoli CA, Selinger DA, Pikaard CS, Richards EJ, Bender J, Mount DW, Jorgensen RA (2002) Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res 30:5036–5055
Perrella G, Lopez-Vernaza MA, Carr C, Sani E, Gossele V, Verduyn C, Kellermeier F, Hannah MA, Amtmann A (2013) Histone deacetylase complex1 expression level titrates plant growth and abscisic acid sensitivity in Arabidopsis. Plant Cell 25:3491–3505
Probst AV, Fagard M, Proux F, Mourrain P, Boutet S, Earley K, Lawrence RJ, Pikaard CS, Murfett J, Furner I, Vaucheret H, Scheid OM (2004) Arabidopsis histone deacetylase HDA6 is required for maintenance of transcriptional gene silencing and determines nuclear organization of rDNA repeats. Plant Cell 16:1021–1034
Sridha S, Wu K (2006) Identification of AtHD2C as a novel regulator of abscisic acid responses in Arabidopsis. Plant J 46:124–133
Tanaka M, Kikuchi A, Kamada H (2008) The Arabidopsis histone deacetylases HDA6 and HDA19 contribute to the repression of embryonic properties after germination. Plant Physiol 146:149–161
Tang Y, Liu M, Gao S, Zhang Z, Zhao X, Zhao C, Zhang F, Chen X (2012) Molecular characterization of novel TaNAC genes in wheat and overexpression of TaNAC2a confers drought tolerance in tobacco. Physiol Plant 144:210–224
Tessadori F, van Zanten M, Pavlova P, Clifton R, Pontvianne F, Snoek LB, Millenaar FF, Schulkes RK, van Driel R, Voesenek LACJ, Spillane C, Pikaard CS, Fransz P, Peeters AJM (2009) PHYTOCHROME B and HISTONE DEACETYLASE 6 control light-induced chromatin compaction in Arabidopsis thaliana. Plos Genet 5:e1000638
Tian L, Chen ZJ (2001) Blocking histone deacetylation in Arabidopsis induces pleiotropic effects on plant gene regulation and development (vol 98, pg 200, 2001). Proc Natl Acad Sci USA 98:7647–7647
Tian L, Wang JL, Fong MP, Chen M, Cao HB, Gelvin SB, Chen ZJ (2003) Genetic control of developmental changes induced by disruption of Arabidopsis histone deacetylase 1 (AtHD1) expression. Genetics 165:399–409
Tian L, Fong MP, Wang JYJ, Wei NE, Jiang HM, Doerge RW, Chen ZJ (2005) Reversible histone acetylation and deacetylation mediate genome-wide, promoter-dependent and locus-specific changes in gene expression during plant development. Genetics 169:337–345
To TK, Kim J-M, Matsui A, Kurihara Y, Morosawa T, Ishida J, Tanaka M, Endo T, Kakutani T, Toyoda T, Kimura H, Yokoyama S, Shinozaki K, Seki M (2011) Arabidopsis HDA6 regulates locus-directed heterochromatin silencing in cooperation with MET1. Plos Genet 7:e1002055
Vrebalov J, Ruezinsky D, Padmanabhan V, White R, Medrano D, Drake R, Schuch W, Giovannoni J (2002) A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (Rin) locus. Science 296:343–346
Wang Z, Cao H, Sun Y, Li X, Chen F, Carles A, Li Y, Ding M, Zhang C, Deng X, Soppe WJJ, Liu Y-X (2013) Arabidopsis paired amphipathic helix proteins SNL1 and SNL2 redundantly regulate primary seed dormancy via abscisic acid-ethylene antagonism mediated by histone deacetylation. Plant Cell 25:149–166
Wang Z, Cao H, Chen F, Liu Y (2014) The roles of histone acetylation in seed performance and plant development. Plant Physiol Biochem 84:125–133
Waterborg JH (2002) Dynamics of histone acetylation in vivo. A function for acetylation turnover? Biochem Cell Biol 80:363–378
Waterborg JH (2011) Plant histone acetylation: in the beginning. Biochimica Et Biophysica Acta 1809:353–359
Wilkinson JQ, Lanahan MB, Yen HC, Giovannoni JJ, Klee HJ (1995) AN ETHYLENE-INDUCIBLE COMPONENT OF SIGNAL-TRANSDUCTION ENCODED BY NEVER-RIPE. Science 270:1807–1809
Xu CR, Liu C, Wang YL, Li LC, Chen WQ, Xu ZH, Bai SN (2005) Histone acetylation affects expression of cellular patterning genes in the Arabidopsis root epidermis. Proc Natl Acad Sci USA 102:14469–14474
Yang XJ, Seto E (2007) HATs and HDACs: from structure, function and regulation to novel strategies for therapy and prevention. Oncogene 26:5310–5318
Zhou CH, Zhang L, Duan J, Miki B, Wu KQ (2005) HISTONE DEACETYLASE19 is involved in jasmonic acid and ethylene signaling of pathogen response in Arabidopsis. Plant Cell 17:1196–1204
Zhou Y, Tan B, Luo M, Li Y, Liu C, Chen C, Yu C-W, Yang S, Dong S, Ruan J, Yuan L, Zhang Z, Zhao L, Li C, Chen H, Cui Y, Wu K, Huang S (2013) HISTONE DEACETYLASE19 interacts with HSL1 and participates in the repression of seed maturation genes in Arabidopsis seedlings. Plant Cell 25:134–148
Acknowledgements
This work was supported by National Natural Science Foundation of China (No. 31572129), and the Natural Science Foundation of Chongqing of China (cstc2015jcyjA80026), and the Fundamental Research Funds for the Central Universities (No. 106112015CDJZR235504).
Author Contributions
JG conducted experiments and wrote the manuscript. ZH and XG conceived and designed research. LZ analyzed data. XY and SZ contributed new reagents and plant materials. GC revised the manuscript. All authors read and approved the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Guo, JE., Hu, Z., Guo, X. et al. Molecular Characterization of Nine Tissue-Specific or Stress-Responsive Genes of Histone Deacetylase in Tomato (Solanum lycopersicum). J Plant Growth Regul 36, 566–577 (2017). https://doi.org/10.1007/s00344-016-9660-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00344-016-9660-8