Abstract
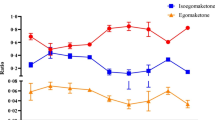
The essential oil of Ocimum species has been extensively used in traditional healthcare in India owing to its pharmacological importance. The popularly renowned ‘Tulsi’ plant is majorly comprised of phytoconstituents belonging to phenylpropanoid and terpenoid classes, albeit a significant variation exists in their proportions in the essential oil. Recent studies from researchers including from our group have defined major genes of phenylpropanoid as well as terpenoid pathways from transcriptomes. Despite transcriptomic resources available, structural information of pathway proteins in Ocimum species for mevalonate pathway (MVA) and 1-deoxy-d-xylulose 5-phosphate (DOXP) terpenoid pathways is limited. The structural information will be useful in defining the differential regulation of pathways in Ocimum species. The present study involved elucidation of molecular structures of proteins in relation to their roles in the control of the MVA pathway in two major species of Ocimum, viz. Ocimum basilicum (Ob) and Ocimum tenuiflorum (Ot). According to transcript analysis, the primary enzymes of the pathway are 3-hydroxy-3-methyl-glutaryl CoA reductase (HMGR) and farnesyl pyrophosphate synthase (FPPS). Structural studies revealed considerable differences in myristoylation and phosphorylation sites of HMGR and FPPS among the two Ocimum species. In both Ocimum species, distinct diversity in helix–helix and β-turns in the secondary structures of HMGR and FPPS proteins resulting in distinct folds, was noted. The presence of aspartate-rich FARM and SARM motif in FPPS was identified through presence of this distinct sequence. Specific amino acid Tyr/Phe at the key position was observed before the FARM motif, suggesting a crucial role in product specificity. Molecular interaction studies and biochemical assays may construe HMGR as a flux regulator for the MVA pathway in Ot. Moreover, docking and enzymatic analysis of FPPS with specific ligands interaction infer diverse product formation. Higher in silico expression supported by quantitative expression of MVA pathway genes in Ot and DOXP pathway gene in Ob showed differential terpenoid biosynthesis. The studies present an insight into the differential regulation of essential oil terpenoid content through participating pathways.




Similar content being viewed by others
Abbreviations
- AACT:
-
Acetoacetyl CoA thiolase
- CMK:
-
4-Cytidine 5′-diphospho-2-C-methyl-d-erythritol kinase
- DMAPP :
-
Dimethylallyl pyrophosphate
- DXR:
-
1-Deoxy-d-xylulose 5-phosphate reductoisomerase
- DXS:
-
1-Deoxy-d-xylulose 5-phosphate synthase
- FPP:
-
Farnesyl pyrophosphate
- FPPS:
-
Farnesyl pyrophosphate synthase
- GPP:
-
Geranyl pyrophosphate
- HMG-CoA:
-
3-Hydroxy-3-methyl glutaryl CoA
- HMGR:
-
3-Hydroxy-3-methyl-glutaryl CoA reductase
- HMGS:
-
3-Hydroxy-3-methyl glutaryl CoA synthase
- IPP:
-
Isopentenyl pyrophosphate
- MEP/DOXP:
-
2-C-methyl-d-erythritol 4-phosphate/1-deoxy-d-xylulose 5-phosphate
- MVA:
-
Mevalonic acid
- MVK:
-
Mevalonate kinase
- Ob :
-
Ocimum basilicum
- Ot :
-
Ocimum tenuiflorum
- PMD:
-
Phosphomevalonate decarboxylase
- PMK:
-
Phosphomevalonate kinase
References
Bansal S, Narnoliya LK, Mishra B, Chandra M, Yadav RK, Sangwan NS (2018) HMG-CoA reductase from Camphor Tulsi (Ocimum kilimandscharicum) regulated MVA dependent biosynthesis of diverse terpenoids in homologous and heterologous plant systems. Sci Rep 8:3547–3561. https://doi.org/10.1038/s41598-017-17153-z
Bayala B, Bassole IH, Scifo R et al (2014) Anticancer activity of essential oils and their chemical components—a review. Am J Cancer Res 4:591–607
Benedict CR, Lu JL, Pettigrew DW et al (2001) The cyclization of farnesyl diphosphate and nerolidyl diphosphate by a purified recombinant delta-cadinene synthase. Plant Physiol 125:1754–1765. https://doi.org/10.1104/pp.125.4.1754
Biasini M, Bienert S, Waterhouse A et al (2014) SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42:252–258. https://doi.org/10.1093/nar/gku340
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. https://doi.org/10.1016/0003-2697(76)90527-3
Ceriotti A, Duranti M, Bollini R (1998) Effects of N-glycosylation on the folding and structure of plant proteins. J Exp Bot 49:1091–1103. https://doi.org/10.1093/jxb/49.324.1091
Chandra M, Kushwaha S, Sangwan NS (2020) Comparative transcriptome analysis to identify putative genes related to trichome development in Ocimum species. Mol Biol Rep 47:6587–6598. https://doi.org/10.1007/s11033-020-05710-1
Chappell J, Wolf F, Proulx J et al (1995) Is the reaction catalyzed by 3-hydroxy-3-methylglutaryl coenzyme a reductase a rate-limiting step for isoprenoid biosynthesis in plants? Plant Physiol 109:1337–1343. https://doi.org/10.1104/pp.109.4.1337
Chaurasiya ND, Sangwan NS, Sabir F, Misra L, Sangwan RS (2012) Withanolide biosynthesis recruits both mevalonate and DOXP pathways of isoprenogenesis in ashwagandha Withania somnifera L. (Dunal). Plant Cell Rep 31:1889–1897. https://doi.org/10.1007/s00299-012-1302-4
Chen D-H, Ye H-C, Li G-F (2000) Expression of a chimeric farnesyl diphosphate synthase gene in Artemisia annua L. transgenic plants via Agrobacterium tumefaciens-mediated transformation. Plant Sci 155:179–185. https://doi.org/10.1016/S0168-9452(00)00217-X
Cheng T, Liu H, Zou H et al (2017) Enzymatic process optimization for the in vitro production of isoprene from mevalonate. Microb Cell Factories 16:8–16. https://doi.org/10.1186/s12934-016-0622-4
Colovos C, Yeates TO (1993) Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci 2:1511–1519. https://doi.org/10.1002/pro.5560020916
Dai Z, Cui G, Zhou SF et al (2011) Cloning and characterization of a novel 3-hydroxy-3-methylglutaryl coenzyme A reductase gene from Salvia miltiorrhiza involved in diterpenoid tanshinone accumulation. J Plant Physiol 168:148–157. https://doi.org/10.1016/j.jplph.2010.06.008
Dale S, Arró M, Becerra B et al (1995) Bacterial expression of the catalytic domain of 3-hydroxy-3-methylglutaryl-coA reductase (isoform HMGR1) from Arabidopsis thaliana, and its inactivation by phosphorylation at Ser577 by Brassica oleracea 3-hydroxy-3-methylglutaryl-coA reducta. Eur J Biochem 233:506–513. https://doi.org/10.1111/j.1432-1033.1995.506_2.x
Darabi M, Masoudi-Nejad A, Nemat-Zadeh G (2012) Bioinformatics study of the 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMGR) gene in Gramineae. Mol Biol Rep 39:8925–8935. https://doi.org/10.1007/s11033-012-1761-2
Daudonnet S, Karst F, Tourte Y (1997) Expression of the farnesyl diphosphate synthase gene of Saccharomyces cerevisiae in tobacco. Mol Breed 3:137–145. https://doi.org/10.1023/a:1009685032495
De Beer TAP, Berka K, Thornton JM, Laskowski RA (2014) PDBsum additions. Nucleic Acids Res 42:292–296. https://doi.org/10.1093/nar/gkt940
Dhar MK, Koul A, Kaul S (2013) Farnesyl pyrophosphate synthase: a key enzyme in isoprenoid biosynthetic pathway and potential molecular target for drug development. N Biotechnol 30:114–123. https://doi.org/10.1016/j.nbt.2012.07.001
Dong N, Ponciano G, McMahan CM et al (2013) Overexpression of 3-hydroxy-3-methylglutaryl coenzyme A reductase in Parthenium argentatum (Guayule). Ind Crops Prod 46:15–24. https://doi.org/10.1016/j.indcrop.2012.12.044
Fraga BM (2013) Natural sesquiterpenoids. Nat Prod Rep 30:1226–1239. https://doi.org/10.1039/c3np70047j
Gorbunov D, Sturlese M, Nies F et al (2014) Molecular architecture and the structural basis for anion interaction in prestin and SLC26 transporters. Nat Commun 5:3622–3635. https://doi.org/10.1038/ncomms4622
Hakkim FL, Arivazhagan G, Boopathy R (2008) Antioxidant property of selected Ocimum species and their secondary metabolite content. J Med Plants Res 2:250–257
Hemmerlin A (2013) Post-translational events and modifications regulating plant enzymes involved in isoprenoid precursor biosynthesis. Plant Sci 203:41–54. https://doi.org/10.1016/j.plantsci.2012.12.008
Hu Y, Gai Y, Yin L et al (2010) Crystal structures of a Populus tomentosa 4-coumarate: CoA ligase shed light on its enzymatic mechanisms. Plant Cell 22:3093–3104. https://doi.org/10.1105/tpc.109.072652
Hu T, Gao ZQ, Hou JM, Tian SK, Zhang ZX, Yang L, Liu Y (2020) Identification of biosynthetic pathways involved in flavonoid production in licorice by RNA-seq based transcriptome analysis. Plant Growth Regul 92(1):15–28
Iijima Y (2004) The biochemical and molecular basis for the divergent patterns in the biosynthesis of terpenes and phenylpropenes in the peltate glands of three cultivars of basil. Plant Physiol 136:3724–3736. https://doi.org/10.1104/pp.104.051318
Jadaun JS, Sangwan NS, Narnoliya LK, Singh N, Bansal S, Mishra B, Sangwan RS (2017) Over-expression of DXS gene enhances terpenoidal secondary metabolite accumulation in rose-scented Geranium and Withania somnifera: active involvement of plastid isoprenogenic pathway in their biosynthesis. Physiol Plant 159:381–400. https://doi.org/10.1111/ppl.12507
Jayaram B, Bhushan K, Shenoy SR et al (2006) Bhageerath: an energy based web enabled computer software suite for limiting the search space of tertiary structures of small globular proteins. Nucleic Acids Res 34:6195–6204. https://doi.org/10.1093/nar/gkl789
Kallberg M, Wang H, Wang S et al (2012) Template-based protein structure modeling using the RaptorX web server. Nat Protoc 7:1511–1522. https://doi.org/10.1038/nprot.2012.085
Kellogg BA, Poulter CD (1997) Chain elongation in the isoprenoid biosynthetic pathway. Curr Opin Chem Biol 1:570–578. https://doi.org/10.1016/S1367-5931(97)80054-3
Kemp LE, Bond CS, Hunter WN (2002) Structure of 2C-methyl-d-erythritol 2,4-cyclodiphosphate synthase: an essential enzyme for isoprenoid biosynthesis and target for antimicrobial drug development. Proc Natl Acad Sci USA 99:6591–6596. https://doi.org/10.1073/pnas.102679799
Khoury GA, Baliban RC, Floudas CA (2011) Proteome-wide post-translational modification statistics: frequency analysis and curation of the Swiss-Prot database. Sci Rep 1:90–105. https://doi.org/10.1038/srep00090
Kim DE, Chivian D, Baker D (2004) Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res 32:526–531. https://doi.org/10.1093/nar/gkh468
Kim Y-J, Lee OR, Oh JY et al (2014) Functional analysis of HMGR encoding genes in triterpene saponin-producing Panax ginseng Meyer. Plant Physiol 165:373–387. https://doi.org/10.1104/pp.113.222596
Kiran U, Ram M, Khan MA et al (2010) Structural and functional characterization of HMG-CoA reductase from Artemisia annua. Bioinformation 5:146–149. https://doi.org/10.6026/97320630005146
Kumar R, Sangwan RS, Mishra S, Sabir F, Sangwan NS (2012) In silico motif diversity analysis of the glycon preferentiality of plant secondary metabolic glycosyltransferases. Plant Omics 5:200–210
Kushwaha AK, Sangwan NS, Trivedi PK, Negi AS, Misra L, Sangwan RS (2013) Tropine forming tropinone reductase gene from Withania somnifera (Ashwagandha): biochemical characteristics of the recombinant enzyme and novel physiological overtones of tissue-wide gene expression patterns. PLoS ONE 8:74777–74789. https://doi.org/10.1371/journal.pone.0074777
Labra M, Miele M, Ledda B et al (2004) Morphological characterization, essential oil composition and DNA genotyping of Ocimum basilicum L. cultivars. Plant Sci 167:725–731. https://doi.org/10.1016/j.plantsci.2004.04.026
Laskowski RA, Rullmannn JA, MacArthur MW et al (1996) AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR 8:477–486. https://doi.org/10.1007/BF00228148
Liang Y, Jiang XM, Hu Q et al (2014) Cloning and characterization of 3-hydroxy-3-methylglutaryl-CoA reductase (HMGR) gene from Paris fargesii Franch. Indian J Biochem Biophys 51:201–206
Lombard J, Moreira D (2011) Origins and early evolution of the mevalonate pathway of isoprenoid biosynthesis in the three domains of life. Mol Biol Evol 28:87–99. https://doi.org/10.1093/molbev/msq177
Maurya S, Sangwan NS (2020) Profiling of essential oil constituents in Ocimum species. Proc Natl Acad Sci India B 90:577–583. https://doi.org/10.1007/s40011-019-01123-8
Maurya S, Chandra M, Yadav RK, Narnoliya LK, Sangwan RS, Bansal S, Sandhu P, Singh U, Kumar D, Sangwan NS (2019) Interspecies comparative features of trichomes in Ocimum reveal insights for biosynthesis of specialized essential oil metabolites. Protoplasma. https://doi.org/10.1007/s00709-018-01338-y
Meigs TE, Roseman DS, Simoni RD (1996) Regulation of 3-hydroxy-3-methylglutaryl-coenzyme A reductase degradation by the nonsterol mevalonate metabolite farnesol in vivo. J Biol Chem 271:7916–7922. https://doi.org/10.1074/jbc.271.14.7916
Miziorko HM (2011) Enzymes of the mevalonate pathway of isoprenoid biosynthesis. Arch Biochem Biophys 505:131–143. https://doi.org/10.1016/j.abb.2010.09.028
Mukherjee P, Desai PV, Srivastava A et al (2008) Probing the structures of leishmanial farnesyl pyrophosphate synthases: homology modeling and docking studies. J Chem Inf Model 48:1026–1040. https://doi.org/10.1021/ci700355z
Mukherjee S, Basu S, Zhang K (2019) Farnesyl pyrophosphate synthase is essential for the promastigote and amastigote stages in Leishmania major. Mol Biochem Parasitol 230:8–15. https://doi.org/10.1016/j.molbiopara.2019.03.001
Narnoliya LK, Rajakani R, Sangwan RS, Gupta V, Sangwan NS (2014) Comparative transcripts profiling of fruit mesocarp and endocarp relevant to secondary metabolism by suppression subtractive hybridization in Azadirachta indica (neem). Mole Biol Rep 41:3147–3162
Narnoliya LK, Kaushal G, Singh SP, Sangwan RS (2017) De novo transcriptome analysis of rose-scented geranium provides insights into the metabolic specificity of terpene and tartaric acid biosynthesis. BMC Genomics 18:74. https://doi.org/10.1186/s12864-016-3437-0
Narnoliya LK, Sangwan N, Jadaun JS, Bansal S, Sangwan RS (2021) Defining the role of a caffeic acid 3-O-methyltransferase from Azadirachta indica fruits in the biosynthesis of ferulic acid through heterologous over-expression in Ocimum species and Withania somnifera. Planta 253:20. https://doi.org/10.1007/s00425-020-03514-y
Needleman SB, Wunsch CD (1970) A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol 48:443–453. https://doi.org/10.1016/0022-2836(70)90057-4
Pagni M, Ioannidis V, Cerutti L et al (2007) MyHits: improvements to an interactive resource for analyzing protein sequences. Nucleic Acids Res 35:W433–W437. https://doi.org/10.1093/nar/gkm352
Pandey G, Madhuri S (2010) Pharmacological activities of Ocimum sanctum (Tulsi): a review. Int J Pharm Sci Rev Res 5:61–66
Pateraki I, Kanellis AK (2010) Stress and developmental responses of terpenoid biosynthetic genes in Cistus creticus subsp. creticus. Plant Cell Rep 29:629–641. https://doi.org/10.1007/s00299-010-0849-1
Pathak R, Narang P, Chandra M et al (2014) Homology modeling and comparative profiling of superoxide dismutase among extremophiles: Exiguobacterium as a model organism. Indian J Microbiol 54:450–458. https://doi.org/10.1007/s12088-014-0482-8
Paton A, Harley RM, Harley MM (1999) Ocimum: an overview of classification and relationships. In: Basil: the genus Ocimum. CRC Press, Boca Raton, pp 1–38
Pattanayak P, Behera P, Das D, Panda S (2010) Ocimum sanctum Linn. A reservoir plant for therapeutic applications: an overview. Pharmacogn Rev 4:95. https://doi.org/10.4103/0973-7847.65323
Pettersen EF, Goddard TD, Huang CC et al (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. https://doi.org/10.1002/jcc.20084
Rastogi S, Meena S, Bhattacharya A, Ghosh S, Shukla RK, Sangwan NS, Lal RK, Gupta MM, Lavania UC, Gupta V, Nagegowda DA (2014) De novo sequencing and comparative analysis of holy and sweet basil transcriptomes. BMC Genomics 15:588–606. https://doi.org/10.1186/1471-2164-15-588
Reuter K, Sanderbrand S, Jomaa H et al (2002) Crystal structure of 1-deoxy-d-xylulose-5-phosphate reductoisomerase, a crucial enzyme in the non-mevalonate pathway of isoprenoid biosynthesis. J Biol Chem 277:5378–5384. https://doi.org/10.1074/jbc.M109500200
Sangwan NS, Farooqi AHA, Shabih F, Sangwan RS (2001) Regulation of essential oil production in plants. Plant Growth Regul 34:3–21. https://doi.org/10.1023/A:1013386921596
Schmidberger JW, Schnell R, Schneider G (2015) Structural characterization of substrate and inhibitor binding to farnesyl pyrophosphate synthase from Pseudomonas aeruginosa. Acta Crystallogr D 71:721–731. https://doi.org/10.1107/S1399004715001121
Sigman L, Sánchez VM, Turjanski AG (2006) Characterization of the farnesyl pyrophosphate synthase of Trypanosoma cruzi by homology modeling and molecular dynamics. J Mol Graph Model 25:345–352. https://doi.org/10.1016/j.jmgm.2006.02.001
Snyder DA, Bhattacharya A, Huang YJ, Montelione GT (2005) Assessing precision and accuracy of protein structures derived from NMR data. Proteins Struct Funct Bioinform 59:655–661. https://doi.org/10.1002/prot.20499
Song AA-L, Abdullah JO, Abdullah MP et al (2012) Overexpressing 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGR) in the lactococcal mevalonate pathway for heterologous plant sesquiterpene production. PLoS ONE 7:52444–52458. https://doi.org/10.1371/journal.pone.0052444
Stermer BA, Bianchini GM, Korth KL (1994) Regulation of HMG-CoA reductase activity in plants. J Lipid Res 35:1133–1140
Tamura K, Stecher G, Peterson D et al (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. https://doi.org/10.1093/molbev/mst197
Tan Q-Q, Zhu L, Li Y et al (2015) A de novo transcriptome and valid reference genes for quantitative real-time PCR in Colaphellus bowringi. PLoS ONE 10:118693–118703. https://doi.org/10.1371/journal.pone.0118693
Tarshis L, Proteau P, Kellogg B et al (1996) Regulation of product chain length by isoprenyl diphosphate synthases. Proc Natl Acad Sci USA 93:15018–15023. https://doi.org/10.1073/pnas.93.26.15018
Thulasiram HV, Poulter CD (2006) Farnesyl diphosphate synthase: the art of compromise between substrate selectivity and stereoselectivity. J Am Chem Soc 128:15819–15823. https://doi.org/10.1021/ja065573b
Tripathi S, Jadaun JS, Chandra M, Sangwan NS (2016) Medicinal plant transcriptomes: the new gateways for accelerated understanding of plant secondary metabolism. Plant Genet Resour 14:256–269. https://doi.org/10.1017/S1479262116000162
Tripathi S, Sangwan RS, Narnoliya LK, Srivastava Y, Mishra B, Sangwan NS (2017) Transcription factor repertoire in Ashwagandha (Withania somnifera) through analytics of transcriptomic resources: insights into regulation of development and withanolide metabolism. Sci Rep 7:16649–16666. https://doi.org/10.1038/s41598-017-14657-6
Tripathi S, Sangwan RS, Mishra B, Jadaun JS, Sangwan NS (2020) Berry transcriptome: insights into a novel resource to understand development dependent secondary metabolism in Withania somnifera (Ashwagandha). Physiol Plant 168:148–173. https://doi.org/10.1111/ppl.12943
Trott O, Olson AJ (2009) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. https://doi.org/10.1002/jcc.21334
Uchida H, Mizohata E, Okada S (2018) Docking analysis of models for 4-hydroxy-3-methylbut-2-enyl diphosphate reductase and a ferredoxin from Botryococcus braunii, race B. Plant Biotechnol 35:297–301. https://doi.org/10.5511/plantbiotechnology.18.0601a
Vishwakarma RK, Patel KA, Sonawane P et al (2012) Molecular characterization of farnesyl pyrophosphate synthase from Bacopa monniera by comparative modeling and docking studies. Bioinformation 8:1075–1081. https://doi.org/10.6026/97320630081075
Vranová E, Coman D, Gruissem W (2012) Structure and dynamics of the isoprenoid pathway network. Mol Plant 5:318–333. https://doi.org/10.1093/mp/sss015
Vranová E, Coman D, Gruissem W (2013) Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu Rev Plant Biol 64:665–700. https://doi.org/10.1146/annurev-arplant-050312-120116
Webb B, Sali A (2014) Protein structure modeling with MODELLER. In: Protein structure prediction. Humana Press, New York, pp 1–15
Zhao N, Ferrer J-L, Ross J et al (2007) Structural, biochemical, and phylogenetic analyses suggest that indole-3-acetic acid methyltransferase is an evolutionarily ancient member of the SABATH family. Plant Physiol 146:455–467. https://doi.org/10.1104/pp.107.110049
Zheng J, Knighton DR, Taylor SS et al (1993) Crystal structures of the myristylated catalytic subunit of cAMP-dependent protein kinase reveal open and closed conformations. Protein Sci 2:1559–1573. https://doi.org/10.1002/pro.5560021003
Acknowledgements
NSS is thankful to Research Grants from CSIR-Network Projects BSC 0203, BSC0107, and HCP007. MC is thankful to UGC for the award of the Fellowship.
Author information
Authors and Affiliations
Contributions
NSS planned the study and experiments. MC performed major experiments, compilation of data, and prepared the draft manuscript. MC, BM and SK are involved in wet lab validation. MC wrote, NSS edited the MS, all authors have gone through the ms.
Corresponding author
Additional information
Communicated by Feibo Wu.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
10725_2022_796_MOESM2_ESM.pptx
Supplementary file2 (PPTX 1277 kb) Supplementary Fig. 1 a Seedlings of O. basilicum and O. tenuiflorum. b Mature plants of O. basilicum and O. tenuiflorum. c Actin PCR amplification of prepared cDNA from different developmental stages of Ocimum species. Supplementary Fig. 2 Predicted 3D structure of MVA pathway enzymes in O. basilicum (Ob) and O. tenuiflorum (Ot). Yellow color highlights the helix in the structures, and strands are marked by purple color and coils are shown by cyan color. PDB ID used for predicting homology models, (PDB 409A) for AACT (acetoacetyl CoA thiolase), (PDB 2F82) for HMGS (3-hydroxy-3-methyl glutaryl CoA synthase), (PDB 1DQA) for HMGR (3-hydroxy-3-methyl glutaryl CoA reductase), (PDB 2R3V) for MVK (mevalonate kinase), (PDB 3D4J) for PMD (diphosphomevalonate decarboxylase), and (PDB 4KK2) for FPPS (farnesyl pyrophosphate synthase). Supplementary Fig. 3 Superimposition of models. The superimposed O. basilicum with O. tenuiflorum with their respective templates. Blue color shows the template, O. basilicum and O. tenuiflorum are shown with brown and pink color. AACT acetoacetyl CoA thiolase, HMGS 3-hydroxy-3-methyl glutaryl CoA synthase, HMGR 3-hydroxy-3-methyl glutaryl CoA reductase, MVK mevalonate kinase, PMK phosphomevalonate kinase, PMD diphosphomevalonate decarboxylase, FPPS farnesyl pyrophosphate synthase. Supplementary Fig. 4 3D representation of binding pockets of FPPS with substrates (IPP and DMAPP). A Ob IPP, B Ob DMAPP, C Ob IPP and DMAPP, D Ot IPP, E Ot DMAPP, and F Ot IPP and DMAPP
Rights and permissions
About this article
Cite this article
Chandra, M., Kushwaha, S., Mishra, B. et al. Molecular and structural insights for the regulation of terpenoids in Ocimum basilicum and Ocimum tenuiflorum. Plant Growth Regul 97, 61–75 (2022). https://doi.org/10.1007/s10725-022-00796-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-022-00796-y