Abstract
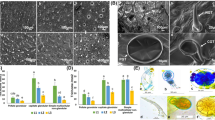
Genus Ocimum is known to have species possessing important therapeutic essential oil. The major phytoconstituents of essential oil in Ocimum species are phenylpropanoids and terpenoids. The essential oil is accumulated in the trichomes; the specialized structures predominantly found on leaves and other tissues. The development of trichome is integrated with development of plant and leaf and also tightly coordinated with the primary and secondary metabolic pathways producing essential oil constituents. In continuation to our studies on elucidating/understanding the mechanism of biosynthesis of essential oil pathways in Ocimum species, we have performed comparative transcriptome analysis to investigate the role of trichome-related gene expression in the regulation of biosynthetic pathways of essential oil. The essential oil biogenesis is tightly integrated with primary metabolic activities, the analysis for the expression pattern of genes related to primary metabolism and its relationship with secondary metabolism was evaluated in comparative manner. Physiological parameters in relation to primary metabolism such as photosynthetic pigment content, soluble sugar content, and invertase enzymes along with morphological parameters were analysed in O. basilicum and O. sanctum. Differential expression profiling uncovered about 8116 and 2810 differentially expressed transcripts in O. basilicum and O. sanctum, respectively. Enrichment of differentially expressed genes were analysed in relation to metabolic pathways, primary metabolism and secondary metabolism. Trichome related genes identified from the Ocimum species vis-à-vis their expression profiles suggested higher expression in O. basilicum. The findings in this study provide interesting insights into the role of trichome-related transcripts in relation to essential oil content in Ocimum species. The study is valuable as this is the first study on revealing the transcripts and their role in trichome development and essential oil biogenesis in two major species of Ocimum.




Similar content being viewed by others
Abbreviations
- Ob:
-
O. basilicum
- Os:
-
O. sanctum
- CPC:
-
Caprice
- ETC1:
-
Enhancer of TRY and CPC1
- ETC2:
-
Enhancer of TRY and CPC2
- ETC3:
-
Enhancer of TRY and CPC3
- GIS:
-
Glabrous inflorescence stems
- GIS2:
-
Glabrous inflorescence stems2
- GL1:
-
Glabrous1
- GL2:
-
Glabra2
- GL3:
-
Glabra3
- SPL3:
-
Squamosa promoter binding protein Like3
- TCL1:
-
Trichomeless1
- TCL2:
-
Trichomeless2
- TRY:
-
Triptychon
- TT8:
-
Transparent testa8
- UPL3:
-
Ubiquitin protein ligase3
- ZFP:
-
Zinc finger protein
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- B2G:
-
Blast2Go
- BLAST:
-
Basic local alignment search tool
References
Paton A, Harley MR, Harley MM (1999) In: Hiltunen R, Holm Y (eds). Basil: the genus Ocimum. Harwood Academic Publishers, Reading, pp 1–38
da Silveira Agostini-Costa T, Vieira R, Bizzo RH, Silveira D, Gimenes MA (2012) Secondary metabolites. In: Dhanarasu E (ed) Chromatography and its applications. InTech, London, pp 131–164
Shih ML, Morgan JA (2020) Metabolic flux analysis of secondary metabolism in plants. Metab Eng Commun 1:10:e00123. https://doi.org/10.1016/j.mec.2020.e00123
Bartwal A, Mall R, Lohani P, Guru SK, Arora S (2013) Role of secondary metabolites and brassinosteroids in plant defense against environmental stresses. J Plant Growth Regul 32(1):216–232. https://doi.org/10.1007/s00344-012-9272-x
Pagare S, Bhatia M, Tripathi N, Pagare S, Bansal YK (2015) Secondary metabolites of plants and their role: overview. Curr Trends Biotechnol Pharm 9(3):293–304
Maurya S, Chandra M, Yadav RK, Narnoliya LK, Sangwan RS, Bansal S, Sandhu P, Singh U, Kumar D, Sangwan NS (2019) Interspecies comparative features of trichomes in Ocimum reveal insights for biosynthesis of specialized essential oil metabolites. Protoplasma. https://doi.org/10.1007/s00709-018-01338-y
Huchelmann A, Boutry M, Hachez C (2017) Plant glandular trichomes: natural cell factories of high biotechnological interest. Plant Physiol 175:00727. https://doi.org/10.1104/pp.17.00727
Oksanen E (2018) Trichomes form an important first line of defence against adverse environment—New evidence for ozone stress mitigation. Plant Cell Environ 41:1497–1499. https://doi.org/10.1111/pce.13187
Prakash P, Gupta N (2005) Therapeutic uses of Ocimum sanctum linn (Tulsi) with a note on eugenol and its pharmacological actions: a short review. Indian J Physiol Pharmacol 49:125–131. https://doi.org/10.7860/JCDR/2014/9122.4629
Pattanayak P, Behera P, Das D, Panda S (2010) Ocimum sanctum Linn. A reservoir plant for therapeutic applications: an overview. Pharmacogn Rev 4:95. https://doi.org/10.4103/0973-7847.65323
Bansal S, Narnoliya LK, Mishra B, Chandra M, Yadav RK, Sangwan NS (2018) HMG-CoA reductase from Camphor Tulsi (Ocimum kilimandscharicum) regulated MVA dependent biosynthesis of diverse terpenoids in homologous and heterologous plant systems. Sci Rep 8:3547–3561. https://doi.org/10.1038/s41598-017-17153-z
Maurya S, Sangwan NS (2019) Profiling of essential oil constituents in Ocimum species. Proc Natl Acad Sci India Sect B Biol Sci 26:1–7. https://doi.org/10.1007/s40011-019-01123-8
Zhao H, Wang X, Zhu D, Cui S, Li X, Cao Y, Ma L (2012) A single amino acid substitution in IIIf subfamily of basic helix-loop-helix transcription factor AtMYC1 leads to trichome and root hair patterning defects by abolishing its interaction with partner proteins in Arabidopsis. J Biol Chem 287:14109–14121. https://doi.org/10.1074/jbc.M111.280735
Gan L, Xia K, Chen J-G, Wang S (2011) Functional characterization of TRICHOMELESS2, a new single-repeat R3 MYB transcription factor in the regulation of trichome patterning in Arabidopsis. BMC Plant Biol 11:176–188. https://doi.org/10.1186/1471-2229-11-176
Liang G, He H, Li Y, Ai Q, Yu D (2014) MYB82 functions in regulation of trichome development in Arabidopsis. J Exp Bot 65:3215–3223. https://doi.org/10.1093/jxb/eru179
Tripathi S, Srivastava Y, Sangwan RS, Sangwan NS (2020) In silico mining and functional analysis of AP2/ERF gene in Withania somnifera. Sci Rep 10:1–12. https://doi.org/10.1038/s41598-020-60090-7
Kirik V, Simon M, Wester K, Schiefelbein J, Hulskamp M (2004) Enhancer of try and CPC 2 (ETC2) reveals redundancy in the region-specific control of trichome development of Arabidopsis. Plant Mol Biol 55:389–398. https://doi.org/10.1007/s11103-004-0893-8
Hung FY, Chen JH, Feng YR, Lai YC, Yang S, Wu K (2020) Arabidopsis JMJ29 is involved in trichome development by regulating the core trichome initiation gene GLABRA3. Plant J. https://doi.org/10.1111/tpj.14858
Sangwan RS, Tripathi S, Singh J, Narnoliya LK, Sangwan NS (2013) De novo sequencing and assembly of Centella asiatica leaf transcriptome for mapping of structural, functional and regulatory genes with special reference to secondary metabolism. Gene 525:58–76. https://doi.org/10.1016/j.gene.2013.04.057
Tripathi S, Sangwan RS, Mishra B, Jadaun JS, Sangwan NS (2019) Berry transcriptome: insights into a novel resource to understand development dependent secondary metabolism in Withania somnifera (Ashwagandha). Physiol Plant. https://doi.org/10.1111/ppl.12943
Marks MD, Wenger JP, Gilding E, Jilk R, Dixon RA (2009) Transcriptome analysis of Arabidopsis wild-type and gl3-sst sim trichomes identifies four additional genes required for trichome development. Mol Plant 2:803–822. https://doi.org/10.1093/mp/ssp037
Tripathi S, Sangwan RS, Narnoliya LK, Srivastava Y, Mishra B, Sangwan NS (2017) Transcription factor repertoire in Ashwagandha (Withania somnifera) through analytics of transcriptomic resources: insights into regulation of development and withanolide metabolism. Sci Rep 7:16649–16666. https://doi.org/10.1038/s41598-017-14657-6
Narnoliya LK, Sangwan RS, Singh SP (2018) Transcriptome mining and in silico structural and functional analysis of ascorbic acid and tartaric acid biosynthesis pathway enzymes in rose-scanted Geranium. Mol Biol Rep 45:315–326. https://doi.org/10.1007/s11033-018-4164-1
Singh N, Luthra R (1988) Sucrose metabolism and essential oil accumulation during lemongrass (Cymbopogon flexuosus Stapf) leaf development. Plant Sci 57:127–133
Farooqi AHA, Bansal RP, Luthra R, Sangwan Neelam S, Sangwan RS (2000) Physiological, biochemical and environmental aspects of essential oil biosynthesis in aromatic grasses (Cymbopogon spp). In: Kumar S et al (eds) Aromatic grass monograph. CIMAP, Lucknow, pp 199–222
Bose SK, Yadav RK, Mishra S, Sangwan RS, Singh AK, Mishra B, Srivastava AK, Sangwan NS (2013) Effect of gibberellic acid and calliterpenone on plant growth attributes, trichomes, essential oil biosynthesis and pathway gene expression in differential manner in Mentha arvensis L. Plant Physiol Biochem 66:150–158
Yadav RK, Sangwan RS, Sabir F, Srivastava AK, Sangwan NS (2014) Effect of prolonged water stress on specialized secondary metabolites, peltate glandular trichomes, and pathway gene expression in Artemisia annua L. Plant Physiol Biochem 74:70–83. https://doi.org/10.1016/j.plaphy.2013.10.02
Yu L, Liu Y, Xu F (2019) Comparative transcriptome analysis reveals significant differences in the regulation of gene expression between hydrogen cyanide- and ethylene-treated Arabidopsis thaliana. BMC Plant Biol 19:1–19. https://doi.org/10.1186/s12870-019-1690-5
Gerstein MB, Rozowsky J, Yan KK, Wang D, Cheng C, Brown JB, Davis CA, Hillier L, Sisu C, Li JJ, Pei B (2014) Comparative analysis of the transcriptome across distant species. Nature 512:445–448. https://doi.org/10.1038/nature13424
Scheible W-R (2004) Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol 136:2483–2499. https://doi.org/10.1104/pp.104.047019
Primmer CR, Papakostas S, Leder EH, Davis MJ, Ragan MA (2013) Annotated genes and nonannotated genomes: cross-species use of gene ontology in ecology and evolution research. Mol Ecol 22:3216–3241. https://doi.org/10.1111/mec.12309
Keeling CI, Weisshaar S, Ralph SG, Jancsik S, Hamberger B, Dullat HK, Bohlmann J (2011) Transcriptome mining, functional characterization, and phylogeny of a large terpene synthase gene family in spruce (Picea spp.). BMC Plant Biol 11:43. https://doi.org/10.1186/1471-2229-11-43
Rastogi S, Meena S, Bhattacharya A, Ghosh S, Shukla RK, Sangwan NS, Lal RK, Gupta MM, Lavania UC, Gupta V, Nagegowda DA, Shasany AK (2014) De novo sequencing and comparative analysis of holy and sweet basil transcriptomes. BMC Genom 15:588. https://doi.org/10.1186/1471-2164-15-588
Winkelblech J, Fan A, Li S-M (2015) Prenyltransferases as key enzymes in primary and secondary metabolism. Appl Microbiol Biotechnol 99:7379–7397. https://doi.org/10.1007/s00253-015-6811-y
Bilgin DD, Zavala JA, Zhu JI, Clough SJ, Ort DR, DeLUCIA EH (2010) Biotic stress globally downregulates photosynthesis genes. Plant Cell Environ 33:1597–1613. https://doi.org/10.1111/j.1365-3040.2010.02167.x
Rojas CM, Senthil-Kumar M, Tzin V, Mysore KS (2014) Regulation of primary plant metabolism during plant-pathogen interactions and its contribution to plant defense. Front Plant Sci 5:1–12. https://doi.org/10.3389/fpls.2014.00017
Sangwan RS, Sangwan NS (2000) Metabolic and molecular analysis of chemotypic diversity in aromatic grasses (Cymbopogon spp.). In: Kumar S et al (eds) Aromatic Grass Monograph. CIMAP, Lucknow, pp 223–247
Sharma PK, Sangwan Neelam S, Mishra BN, Sangwan RS (2009) Coherent ontogenic dynamics of geraniol acetyltransferase activity and geranyl acetate concentration in flowers and leaves of aroma grass Cymbopogon martini var Motia. Plant Growth Regul 57:103–10851
Sharma P, Sangwan Neelam S, Bose SK, Sangwan RS (2013) Biochemical characteristics of a novel vegetative tissue geraniol acetyltransferase from a monoterpene oil grass (Palmarosa, Cymbopogon martinii var. Motia) leaf. Plant Sci 203–204: 63–73
Ng WL, Wu W, Zou P, Zhou R (2019) Comparative transcriptomics sheds light on differential adaptation and species diversification between two Melastoma species and their F1 hybrid. AoB Plants. https://doi.org/10.1093/aobpla/plz019
Fasbender L, Yáñez-Serrano AM, Kreuzwieser J, Dubbert D, Werner C (2018) Real-time carbon allocation into biogenic volatile organic compounds (BVOCs) and respiratory carbon dioxide (CO2) traced by PTR-TOF-MS, 13 CO2 laser spectroscopy and 13 C-pyruvate labelling. PLoS ONE 13:1–22. https://doi.org/10.1371/journal.pone.0204398
Delfin JC, Watanabe M, Tohge T (2019) Understanding the function and regulation of plant secondary metabolism through metabolomics approaches. Theor Exp Plant Physiol 31:127–138. https://doi.org/10.1007/s40626-018-0126-1
Wagner GJ (1991) Secreting glandular trichomes: more than just hairs. Plant Physiol 96:675–679. https://doi.org/10.1104/pp.96.3.675
Pattanaik S, Patra B, Singh SK, Yuan L (2014) An overview of the gene regulatory network controlling trichome development in the model plant, Arabidopsis. Front Plant Sci 5:1–8. https://doi.org/10.3389/fpls.2014.00259
Gan Y (2006) Glaborous inflorescense stem modulates the regulation by gibberellins of epidermal differentiation and shoot maturation in Arabidopsis. Plant Cell Online 18:1383–1395. https://doi.org/10.1105/tpc.106.041533
Liu CH, Di YP (2020) Analysis of RNA sequencing data using CLC Genomics Workbench. In: Keohavong P, Grant SG (eds) Molecular toxicology protocols. Humana, New York, pp 61–113
Bray NL, Pimentel H, Melsted P, Pachter L (2016) Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34:525–527. https://doi.org/10.1038/nbt.3519
Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, MacManes MD (2013) De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc 8:1494–1512. https://doi.org/10.1038/nprot.2013.084
Babicki S, Arndt D, Marcu A et al (2016) Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res 44:W147–W153. https://doi.org/10.1093/nar/gkw419
Acknowledgements
NSS is thankful to networking projects BSC-0203, and BSC0107 for financial grant from CSIR, N Delhi. MC is thankful to UGC for Junior and senior research fellowship and CIMAP-JNU-PhD program for registration.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Authors declare no conflict of interest; all the co-authors are well aware of the publication of this work. Research not involved Human participants and/or Animals.
Research involving human and animal rights
Research not involved Human participants and/or Animals.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Chandra, M., Kushwaha, S. & Sangwan, N.S. Comparative transcriptome analysis to identify putative genes related to trichome development in Ocimum species. Mol Biol Rep 47, 6587–6598 (2020). https://doi.org/10.1007/s11033-020-05710-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-020-05710-1