Abstract
Salinity is one of the major abiotic stresses limiting rice production worldwide. Understanding the genetic basis of salinity tolerance is key for rice breeding. In this study, a recombinant inbred line (RIL) population derived from a super hybrid rice Liang–You–Pei–Jiu (LYP9) parents 93-11 and PA64s, exhibited variation in phenotypes including shoot length (SL), root length (RL), shoot fresh weight (SFW), root fresh weight (RFW), shoot dry weight (SDW), and root dry weight (RDW) under 50 and 100 mM NaCl stress. QTL analysis identified a total of 38 QTLs for these 6 traits under two distinct concentrations of salt, distributing on chromosomes 1, 2, 3, 4, 5, 6, 7 and 10. A total of 21 QTLs were found in 6 stable loci. A novel major QTL, qSL7 for shoot length on chromosome 7 was identified in two distinct concentrations. A chromosome segment substitution line (CSSL) harboring the qSL7 locus from PA64s with 93-11 background was developed and exhibited higher SL value, higher K+ concentration, and lower Na+ concentration compared to 93-11. With BC5F2:3 derived from CSSL-qSL7/93-11, the qSL7 was fine mapped within a 252.9 kb region on chromosome 7 where 40 annotated genes located including, LOC_Os07g43530, which encodes a DNA-binding domain containing protein reported previously as a transcription factor playing a positive role in salt stress tolerance. Our study provides new genetic resources for improvement of salinity tolerance in rice breeding.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The soil salinity is the critical abiotic stress that limiting crop production all over the world. It is the major problem of rice based farming systems in the coastal areas. Also, inland is affected by salinity due to improper irrigation and drainage system. Approximately 20% irrigated and 8% of rainfed agricultural land is affected by salinity (Asif et al. 2018). About one-third of the irrigated rice growing areas are affected by salinity (Prasad et al. 2000). Salinity stress impacts on rice may vary depending upon its different growth and development stages (Yamaguchi and Blumwald 2005). Rice can tolerate salt stress at late vegetative and maturity stages and susceptible at early the vegetative stage. In general, germination and seedling are the two most sensitive stages of salinity. Salinity tolerance in rice is a physiologically complex trait and attributed to multiple mechanisms. The effect of salinity stress arises as a result of the relationship between the physiological and molecular responses of plants (Zhang et al. 2018). Understanding the salt stress tolerance mechanism of rice is crucial for identifying the responsible genetic material. Progress in rice breeding with salt tolerance is still slow because of genetic complexity (Flowers and Flowers 2005). Besides, salt tolerance screening is difficult because the phenotypic response of rice to salt stress is highly affected by other confounding environmental factors (Flowers 2004). The genetics behind salinity tolerance governed by many genes exhibiting the quantitative or polygenic nature of the trait can be revealed by using quantitative trait locus (QTLs) analysis (Gimhani et al. 2016; Negrão et al. 2011). Mapping of QTLs by using molecular markers can aid the genetic dissection of the salt tolerant genes. Since the advent of molecular markers, QTL analyses for salinity tolerance were conducted using different populations and most of them reported salinity tolerance is a polygenetic trait (De Leon et al. 2016; Gregorio et al. 2002). Therefore, uncovering the genes behind salt tolerant traits remains difficult and vital for the marker-assisted plant breeding approach.
There were lots of studies have been done on salt tolerant QTL mapping but due to lack of enough molecular marker coverage the genomic interval was so large to identify candidate genes like, previously the promising QTL designated as Saltol was mapped in large genomic region of 10.7–12.2 Mb on the short arm of the chromosome 1 (Bonilla et al. 2002; Gregorio et al. 1997). Recently advanced high-density molecular marker technology has been developed for rapid high-throughput genotyping. High-throughput SNP genotyping facilities a number of advantages over previous marker systems providing sufficiently dense genome coverage for the dissection of complex traits (Gimhani et al. 2016; Takehisa et al. 2004; Thomson et al. 2011). Bimpong et al. (2013) mapped salt tolerant QTL by using 384-plex SNP markers. More recently De Leon (De Leon et al. 2016) used 9303 SNP markers generated by genotyping-by-sequencing (GBS) were mapped to 2817 recombination points for salt tolerant QTL identification. In addition, several studies of salinity-tolerant QTLs mapping has been reported although few of them were fine mapped. The large chromosomal intervals delimited the attribution of those QTLs. Finally, the identification of candidate genes become more challenging.
Fine mapping is one of the commonly used approaches to identifying genes underlying the quantitative traits. Number of QTLs have been identified for salt stress related traits in the past among them only qSKC1 was successfully isolated by map based cloning (Ren et al. 2005) and others need to be cloned.
In this study recombinant inbred lines developed from the cross between 93-11 and PA64s were used for QTL analysis. Parent 93-11 is an indica variety widely grown in China and PA64s is a thermosensitive indica variety. A high-density SNP linkage map was used for identification of QTLs associated with 6 important traits under salt stress. A novel QTL, qSL7 for shoot length was fine mapped and delimited into a 252.9 kb region on chromosome 7, which will be helpful for further cloning of it and understanding the genetic basis of rice salinity tolerance.
Materials and methods
Plant materials and growing conditions
A total of 132 RILs derived from the cross between super hybrid rice LYP9’s parents 93-11 and PA64s were used in this study for salinity tolerance screening. Susceptible parent 93-11 is an elite indica variety and tolerant parent PA64s a thermosensitive indica variety. Phenotyping for salt tolerance parameters was commenced on RILs along with two parents (Gao et al. 2013).
Phenotypic evaluation of seedling under salt stress
The parents and RILs were assessed at seedling stage by measuring 6 salinity responsive traits viz. SL, RL, SFW, RFW, SDW, and RDW. The treatments were comprised of three NaCl levels: 0 (control, CK), 50 mM NaCl (moderate salt stress) and 100 mM NaCl (high salt stress). To grow seedlings under hydroponic culture, the grains were surface-sterilized and allowed to germinate in water for 3 days. The germinated seedlings were transferred to nutrient solution containing 1.425 mM NH4NO3, 0.42 mM NaH2PO4, 0.510 mM K2SO4, 0.998 mM CaCl2, 1.643 mM MgSO4, 0.168 mM Na2SiO3, 0.125 mM Fe-EDTA, 0.019 mM H3BO3, 0.009 mM MnCl2, 0.155 mM CuSO4, 0.152 mM ZnSO4, and 0.075 mM Na2MoO4 (Li et al. 2016) and exposed to a 14 h photoperiod, under a temperature regime of 30ºC/25ºC and a relative humidity level of − 70%. The pH was adjusted to 5.5–5.8. The nutrient solution was replaced every 2 days interval to adjust the volume and pH of nutrient solution. At the 7th day after planting, the solution was salinized by the addition of NaCl, to which the seedlings were exposed for 2 weeks. The whole experiment was conducted in randomized complete block design with two biological replications. Fifteen plants per line of uniform growth were evaluated for traits related to salinity tolerance. Shoot length and root length were measured in centimetre. Shoot length was measured from the base of the culm to the tip of the tallest leaf and root length was measured from the base of the culm to the tip of the longest root. For dry weight, five plants per line per replication were collected and dried at 65ºC oven for 5 days prior to weighing.
Na+/K+ Ion Measurement
For the determination of sodium (Na+) and potassium (K+) concentration, 28 days after salinization, the shoot was collected from 93-11 and CSSL-qSL7 (three plants from each genotype). After three times distilled water washing samples were dried at 70 °C for 3 days. Further, 10 mg fine grounded sample was extracted in 10 ml of 0.1 N acetic acid then set in a 90 °C water bath for at least 2 h until the sample was completely digested. Extracted solution was cooled at room temperature and filtered using Whatman 1 filter paper and diluted ten times. Na+ and K+ were measured by flame emission spectrometry using a flame photometer at 589 nm (Sherwood flame photometer, Model 420, Cambridge, UK).
DNA isolation and PCR analysis
The parents and BC5F2 individuals’ genomic DNA was extracted from fresh leaves using the CTAB method described by Luo et al. (2001). The PCR protocol was performed in a 15 μL reaction mix including 25 ng genomic DNA, 2 μL of each primer, 1.0 μL 10 × PCR buffer, 0.1 mmol/L dNTP, 0.2 μL 5 U/μL Taq DNA polymerase (Tiangen Biotech, Beijing, China) and 1.5 μL ddH2O. Amplification conditions consisted of an initial denaturation at 94 °C for 5 min, 40 cycles of 94 °C for 30 s, 55–60 °C for 30 s, and 72 °C for 30 s. The reaction were completed with a final extension at 72 °C for 10 min, and saving at 15 °C forever.
Genetic map construction and QTL analysis
The SNP map covered a total of 1381.9 cM of rice genome (Gao et al. 2013). QTL analysis was conducted with the MultiQTL package (www.multiqtl.com) using the maximum likelihood interval mapping approach for the RILs. The Logarithm of the odds (LOD) threshold and major effect QTLs were obtained based on a permutation test (1000 permutations, P = 0.05) for each dataset. We followed the suggestions by McCouch for the QTL nomenclature (McCouch 2008). New SSR and Indel markers in the targeted genomic region were designed according to genome sequence differences between the indica cv. 93-11 and the indica cv. PA64s. The sequences of these newly developed markers are listed in Table 1.
Construction of fine mapping population
To develop the chromosome segment substitution line (CSSL), a RIL containing PA64s genotype at the qSL7 flanking region on chromosome 7 was selected to backcross with recurrent parent 93-11. Then, a CSSL at the qSL7 locus, CSSL-qSL7 was established and crossed with 93-11. A total of 1300 BC5F2:3 seedlings were evaluated under high (100 mM NaCl) salt stress as above mentioned method to fine map of qSL7 with 8 markers.
Statistical analysis of data
Mean phenotypic values for seedling root traits were compared using the Student’s t test. Phenotypic correlations were calculated using a generalized linear model implemented within the SAS (Statistical Analysis System) v8.01.
Results
Evaluation of phenotypic characteristics under salt stress
The parents and the RILs population were evaluated under control/0 mML−1 (L0), moderate/50 mM (L50) and high/100 mM (L100) NaCl salt stress for SL, RL, SFW, SDW, RFW, and RDW. There was no significant difference in all traits except RFW and RDW between two parents under control (0 mM) condition (Fig. 1 a, d–i). Therefore, the treatment of 0 mM NaCl was not conducted for further QTL analysis. Further, the parents and RILs showed varying levels of tolerance in both (L50 and L100) stress levels. Finally, 50 and 100 mM NaCl treatment was conducted for further study among RILs to fully demonstrate their phenotypic variance. The phenotypic measurement showed that the frequency distributions of all traits in RIL populations accorded with the normal distribution and the acceptable range of skewness below + 1.5 and above − 1.5 (Table 2).
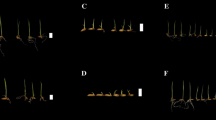
Phenotypes comparison between two parental lines. a–c Morphological responses of 93-11 and PA64s under 0, 50 and 100 mM NaCl stress, respectively; 93-11 is in left and PA64s in right side. d–iSL shoot length, RL root length, SFW shoot fresh weight, SDW shoot dry weight, RFW root fresh weight and RDW root dry weight of parents 93-11 and PA64s. Data represent the mean ± SD of two biological replicates (Student’s t test: *P < 0.05, **P < 0.01) respectively
At both stress conditions (L50 and L100), two parents 93-11 and PA64s showed significant different responses in all traits (Fig. 1b–c, d–i; Table 2). PA64s showed consistently higher SL, RL, SFW, SDW, RFW, and RDW than 93-11 in both stress levels. Among the RILs, differences were found for all traits, indicating a wide range of variation and the mean values of RILs range between the parental means for all 6 traits. As indicated in the frequency distribution (Fig. 2) and the range of RIL values for each trait (Table 2), several lines showed phenotypically superior to the parents. The six traits in the RIL population exhibited different degrees of transgressive segregation. The results indicated that the recurrent parent 93-11 was more susceptible to salt stress and that the PA64s genome might harbour beneficial alleles associated with enhanced salt tolerance.
Correlation analysis of six traits
The correlation analysis showed that there were positive significant correlations among SL, RL, SFW, SDW, RFW, and RDW under two different salinity conditions (Table 3). According to the correlation analysis, the SL is significantly correlated with RL, SFW, SDW, RWF, and RDW. Although, there was no significant correlation between RL and SFW, and SL and RDW in L50 where coefficient were 0.11 and 0.16, respectively. Moreover, SL was highly significant with all the traits in L100 level. Thus, the association among all morphological traits with each other indicated the co-dependence of phenotypic responses for salinity tolerance and presence of QTL clusters in those regions.
Identification of QTLs salt tolerance traits at seedling stage
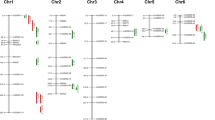
A total of 38 QTLs affecting SL, RL, SFW, SDW, RFW, and RDW were detected across two stress levels and were distributed on chromosomes 1, 2, 3, 4, 5, 6, 7 and 10 (Table 4, Fig. 3). QTLs, with either negative or positive additive effect were found, which indicated that both parents contributed favorable alleles. More favourable alleles come from tolerant parent PA64s. Ten QTLs were associated with SL detected on chromosomes 1, 3, 6, 7 and 10 and each accounted for 6.8–18.20% of the phenotypic variation with LOD values ranging from 2.6 to 5.9. Eleven QTLs for RL were identified on chromosomes 2, 3, 4, 5, 6, 9 and 10 and each accounted for 5.9–23.7% of phenotypic variation. Five QTLs related to SFW on chromosome 1, 6 and 7, each explained 9.8–18.8% phenotypic variation. Six QTLs associated with SDW were detected on chromosomes 6, 7 and 10 were found with 11.2–16.3% phenotypic variation and LOD values ranges from 3.23 to 4.93. Only two QTLs detected on chromosomes 1 and 6 associated with RFW and each explained 7.50% and 10.8% phenotypic variation. Four QTLs were identified for RDW on chromosomes 3, 6 and 7 and each associated with 9.8–12.1% phenotypic variation.
Six QTL loci with five QTLs clusters were repeatedly detected across the two stress levels on chromosomes 1, 3, 6 and 7 (Fig. 3). Thus, 21 QTLs were identified as stable QTLs (Table 5). Stable QTLs were defined based on three criteria, including overlapped QTLs associated with multiple traits in a single stress level, QTLs repeatedly detected in the two stress levels and locus found overlapped with previously reported QTLs. Except for locus 4, all loci of cluster were found overlapped with previous studies (Bimpong et al. 2013; De Leon et al. 2016; Ghomi et al. 2013; Kim et al. 2009; Kumar et al. 2015; Lekklar et al. 2019). Locus 4 consisted of a novel QTL qSL7 (L50 and L100), and explained 7.5% (L50) and 6.8% (L100) phenotypic variation (Table 4).
Development and characterization of CSSL-qSL7 under salinity
In order to confirm the effect of qSL7 for seedling height under salt stress, one chromosomal segment substitution line (CSSL) CSSL-qSL7, harboring the segment of qSL7 from PA64s between the two SSR markers RM21930 and RM22105 (Fig. 4c, d), was developed. The parent 93-11 and CSSL-qSL7 were evaluated under control and salt stress conditions (Fig. 4a, b), the seedling height of the CSSL-qSL7 was significantly higher than that of 93-11 (Fig. 4e). Ion analysis showed that the concentration of K+ and Na+ were significantly higher and lower, respectively in tolerant CSSL-qSL7 than 93-11 (Fig. 4f, g), which indicated CSSL-qSL7 was significantly tolerant to salt stress than 93-11. Therefore, the CSSL-qSL7 was backcrossed with the parent 93-11 and a total of 1300 (BC5F2:3) seedlings obtained from the cross were screened for fine mapping of qSL7 under salt stress.
Phenotypic and genotypic performance of the parent and CSSL-qSL7. a, b Shoot morphology of 93-11 (left) and CSSL-qSL7 (right) under control and stress condition. c Graphical genotype of CSSL-qSL7 compared to the parental lines (chromosomal segments derived from PA64s in black). d Genetic location of qSL7 on chromosome 7. e–g Shoot length, shoot K+ and Na+ concentration of 93-11 and CSSL-qSL7, respectively
Fine mapping of qSL7
For fine mapping, a total of 1300 BC5F2:3 individual plants derived from CSSL-qSL7/93-11 were screened with SSR markers RM21930 and RM22105 flanking qSL7. Based on the resequencing data of 93-11 and PA64s, several Insertion/Deletion markers were developed to screen the population (Table 1). According to the SL value, only homozygous recombinant lines with SL > 24 cm or SL < 20 cm were selected for fine mapping. Combined phenotype and genotype analysis, qSL7 was finally delimited to a 252.9 kb region between markers Indel 7-2 and Indel 7-3 on chromosome 7 (Fig. 5). According to the Rice Genome Annotation Project Database (http://rice.plantbiology.msu.edu/cgi-bin/gbrowse/rice/), there were 40 annotated genes in the target region (Table 6), including 33 expressed proteins, a hypothetical protein, and 6 retrotransposons. Among them, LOC_Os07g43530, which encodes the DNA-binding domain-containing protein, was reported previously as a major transcription factor regulating genes for potassium ion transporters and thereby controlling potassium homeostasis during salt stress.
Discussion
Advancement in the salt tolerant rice breeding improvement would have a significant impact on food security particularly when the world facing the sea level rising issue due to global warming. However, developing tolerant varieties requires sufficient genetic diversity. QTL mapping is one of the effective ways of breeding programs to discover underlying salt tolerant genes of quantitative traits. Previously many QTLs related to salt tolerance have been identified most of them covered large chromosomal intervals due to less availability of polymorphic marker, low-throughput, and the uneven genome-wide distribution (De Leon et al. 2016; Gimhani et al. 2016). However, the breakthrough of modern technology in molecular sector facilitated the high-resolution QTL mapping by the implementation of the high-throughput SNP linkage map. In this study, we utilized a high-throughput genetic linkage map of a RIL population derived from a cross between 93-11 and PA64s to identify QTLs for SL, RL, SFW, SDW, RFW and RDW under moderate and high salt stress at the seedling stage. Rice shows sensitivity to salt stress mostly at the seedling stage of crop development with a threshold as low as an EC of − 3 dSm−1 (Ahn et al. 2004; Bimpong et al. 2013). Salt stress resulted in a substantial reduction in root and shoot length followed by decreased fresh and dry weight in RILs across both stress conditions compared to the control condition. Present findings revealed 38 putative QTLs for 6 traits on chromosomes 1,2,3,4,5,6,7 and 10.
Several studies have been reported that usually QTL cluster responsible for correlated traits lie in the same genomic region confirming stable QTL (Gimhani et al. 2016; Marathi et al. 2012; Yang et al. 2019). This QTLs cluster of different traits may be due to the presence of pleiotropy or tight linkage of genes controlling the traits (Gao et al. 2013; Gimhani et al. 2016). Therefore, co-located chromosomal regions responsible for different salinity tolerant traits may have an opportunity to introgress them together as a unit into rice varieties through MAS/MAB and to develop resilient salt-tolerant cultivars (Gimhani et al. 2016). In this study, the traits are significantly correlated to each other, confirming that QTLs responsible for different traits were co-localized at the specific genomic regions across chromosomes 1, 3, 6 and 7 forming strong QTL clusters. Six stable loci with five QTL clusters associated with salt tolerance were obtained, and each QTL accounted for 6.8–18.8% of the corresponding phenotypic variation. Further, cluster 1, 2 and 3 contain 4, 3 and 5 QTLs, respectively and locus 4 and 5 contains 2 QTLs each. Cluster 1 contains 4 QTLs across both stress levels which affects SL, SFW and RFW shared a similar chromosomal region with previously identified QTL in between marker S1_5501756 and S1_5792183 by De Leon et al. (2016) using the SNP linkage map with indica derived mapping population. Also, a QTL cluster was found in the same location with cluster 1 in salt tolerant introgression lines in rice (Zang et al. 2008). In cluster 3 between SNP7-131 and SNP7-170 markers, 5 co-localized QTLs were found for 4 traits (SL, SFW, SDW, and RDW), which is also overlapped with reported QTLs for salt injury score and root length (De Leon et al. 2016; Sabouri et al. 2009). Moreover, in cluster 5 we identified 1 QTL cluster of 2 traits (RL and RDW) under moderate salt stress although it could not withstand under high salt stress level. These results suggested that root is highly affected by high salt concentration in the field. A similar finding was showed by Gimhani et al. (2016). The cluster 6 contains 7 stable QTLs in between marker SNP6-45–SNP6-81 is overlapped with previously reported marker S6_5848568—S6_5905669, RM10-RM234, RM3027-RM5371 and RM586-RM253 (Bimpong et al. 2013; De Leon et al. 2016; Sabouri et al. 2009; Sabouri and Sabouri 2008; Zang et al. 2008).
One major potential QTL in cluster 4, qSL7 accounted for 7.5 and 6.8% phenotypic variation, respectively and the allele contributed from tolerant parents PA64s was identified in both moderate and high salt stress conditions. This locus was stable and not previously reported for shoot length under salt stress condition thus, we selected qSL7 to narrow down further.
Furthermore, one CSSL, CSSL-qSL7, which produced significantly higher shoot length was selected for further study and crossed with the parent 93-11 and a total of 1300 (BC5F2:3) seedlings obtained from the cross were screened for fine mapping of qSL7 under high salt stress condition. Besides, CSSL-qSL7 harboring qSL7 locus was performed significantly better than 93-11 in terms of shoot length and Na/K ion accumulation under high salt stress level, indicating qSL7 could withstand under high salt stress condition. The gene qSL7 was finally fine mapped to a region of 252.9 kb on the long arm of rice chromosome 7. Among 40 predicted genes in this region, only one gene LOC_Os07g43530 DNA-binding domain containing protein belongs to basic helix-loop-helix (bHLH) family has been reported previously as a transcription factor that plays a positive role in salt stress tolerance by binds to the promoter of a potassium ion transporter gene to regulate potassium homeostasis during salt stress (Wu et al. 2015). A number of transcription factor of the bHLH gene family are important for regulating plant growth and plant responses to salt stress, such as OsbHLH094, OsbHLH062, and OsbHLH035 already reported to take part in salt tolerance by DNA-binding in the promoter region of some ion transporter genes (Blumwald 2000; Chen et al. 2018; Toda et al. 2013). Hence, LOC_Os07g43530 may be the candidate gene for the qSL7 locus associated with shoot length.
Conclusion
In this study, an indica derived population with a high-density genetic map was used to identify QTLs for rice salt tolerance at the seedling stage. A total of 38 QTLs were detected across two stress levels. Six cluster were identified with 21 stable QTLs. A novel major QTL for SL, qSL7 was identified on chromosome 7. With the BC5F2:3 population derived from the CSSL-qSL7, it was delimited to a 252.9 kb region. Among 40 annotated genes identified in the target region, LOC_Os07g43530 was previously reported as a transcription factor of the potassium ion transporter gene and might play an important role in regulating shoot length of rice under salt stress. Further study is needed for cloning of qSL7 and understanding of its molecular and biological functions. Finally, these recombinant lines derived from CSSL-qSL7 will provide genetic resources for improvement of rice salinity tolerance.
References
Ahn SJ, Shin R, Schachtman DP (2004) Expression of KT/KUP genes in Arabidopsis and the role of root hairs in K+ uptake. Plant Physiol 134:1135–1145
Asif MA, Schilling RK, Tilbrook J, Brien C, Dowling K, Rabie H, Short L, Trittermann C, Garcia A, Barrett-Lennard EG, Berger B, Mather DE, Gilliham M, Fleury D, Tester M, Roy SJ, Person AS (2018) Mapping of novel salt tolerance QTL in an Excalibur x Kukri doubled haploid wheat population. Theor Appl Genet 131:2179–2196
Bimpong IK, Manneh B, El-Namaky R, Diaw F, Amoah NKA, Sanneh B, Ghislain K, Sow A, Singh R, Gregorio GB, Bizimana JB, Wopereis MC (2013) Mapping QTLs related to salt tolerancein riceatthe young seedling stage using 384-plex single nucleotide polymorphism SNP marker sets. Mol Plant Breed 5:47–63
Blumwald E (2000) Sodium transport and salt tolerance in plants. Curr Opin Cell Biol 12:431–434
Bonilla P, Mackell D, Deal K, Gregorio G (2002) RFLP and SSLP mapping of salinity tolerance genes in chromosome 1 of rice (Oryza sativa L.) using recombinant inbred lines. Philipp Agric Sci 85:68–76
Chen HC, Cheng WH, Hong CY, Chang YS, Chang MC (2018) The transcription factor OsbHLH035 mediates seed germination and enables seedling recovery from salt stress through ABA-dependent and ABA-independent pathways, respectively. Rice 11(1):50
De Leon TB, Linscombe S, Subudhi PK (2016) Molecular dissection of seedling salinity tolerance in rice (Oryza sativa L.) using a high-density GBS-based SNP linkage map. Rice 9:52
Flowers TJ (2004) Improving crop salt tolerance. J Exp Bot 55:307–319
Flowers TJ, Flowers SA (2005) Why does salinity pose such a difficult problem for plant breeders? Agric Water Manag 78:15–24
Gao ZY, Zhao SC, He WM, Guo LB, Peng YL, Wang JJ, Guo XS, Zhang XM, Rao YC, Zhang C, Dong GJ, Zheng FY, Lu CX, Hu J, Zhou Q, Liu HJ, Wu HY, Xu J, Ni PX, Zeng DL, Liu DH, Tian P, Gong LH, Ye C, Zhang GH, Wang J, Tian FK, Xue DW, Liao Y, Zhu L, Chen MS, Li JY, Cheng SH, Zhang GY, Wang J, Qian Q (2013) Dissecting yield-associated locus in super hybrid rice by resequencing recombinant inbred lines and improving parental genome sequences. Proc Natl Acad Sci 110:14492–14497
Ghomi K, Rabiei B, Sabouri H, Sabouri A (2013) Mapping QTLs for traits related to salinity tolerance at seedling stage of rice (Oryza sativa L.): an agrigenomics study of an Iranian rice population. OMICS 17:242–251
Gimhani DR, Gregorio GB, Kottearachchi NS, Samarasinghe WL (2016) SNP-based discovery of salinity-tolerant QTLs in a bi-parental population of rice (Oryza sativa). Mol Genet Genomics 291:2081–2099
Gregorio GB, Senadhira D, Mendoza RD (1997) Screening rice for salinity tolerance. IRRI Discussion paper series no. 22
Gregorio GB, Senadhira D, Mendoza RD, Manigbas NL, Roxas JP, Guerta CQ (2002) Progress in breeding for salinity tolerance and associated abiotic stresses in rice. Field Crops Res 76:91–101
Kim DM, Ju HG, Kwon TR, Oh CS, Ahn SN (2009) Mapping QTLs for salt tolerance in an introgression line population between Japonica cultivars in rice. J Crop Sci Biotechnol 12:121–128
Kumar V, Singh A, Mithra SV, Krishnamurthy SL, Parida SK, Jain S, Tiwari KK, Kumar P, Rao AR, Sharma SK, Khurana JP, Singh NK, Mohapatra T (2015) Genome-wide association mapping of salinity tolerance in rice (Oryza sativa). DNA Res 22:133–145
Lekklar C, Pongpanich M, Suriya-Arunroj D, Chinpongpanich A, Tsai H, Comai L, Chadchawan S, Buaboocha T (2019) Genome-wide association study for salinity tolerance at the flowering stage in a panel of rice accessions from Thailand. BMC Genomics 20:76
Li Q, Yang A, Zhang WH (2016) Efficient acquisition of iron confers greater tolerance to saline-alkaline stress in rice (Oryza sativa L.). J Exp Bot 67:6431–6444
Luo ZY, Zhou G, Chen XH, Lu QH, Hu WX (2001) Isolation of high-quality genomic DNA from plants. Hunan Yi Ke Da Xue Bao 26:178–180
Marathi B, Guleria S, Mohapatra T, Parsad R, Mariappan N, Kurungara VK, Atwal SS, Prabhu KV, Singh NK, Singh AK (2012) QTL analysis of novel genomic regions associated with yield and yield related traits in new plant type based recombinant inbred lines of rice (Oryza sativa L). BMC Plant Biol 12:137
McCouch SR (2008) Gene nomenclature system for rice. Rice 1:72–84
Negrão S, Courtois B, Ahmadi N, Abreu I, Saibo N, Oliveira MM (2011) Recent updates on salinity stress in rice: from physiological to molecular responses. Crit Rev Plant Sci 30:329–377
Prasad SR, Bagali PG, Hittalmani S, Shashidhar HE (2000) Molecular mapping of quantitative trait locus associated with seedling tolerance to salt stress in rice (Oryza sativa L.). Curr Sci 78:162–164
Ren ZH, Gao JP, Li GL, Cai XL, Huang W, Chao DY, Zhu MZ, Wang ZY, Luan S, Lin HX (2005) A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat Genet 37:1141–1146
Sabouri H, Sabouri A (2008) New evidence of QTLs attributed to salinity tolerance in rice. Afr J Biotechnol 7:4376–4383
Sabouri H, Rezai AM, Moumeni A, Kavousi A, Katouzi M, Sabouri A (2009) QTLs mapping of physiological traits related to salt tolerance in young rice seedlings. Biol Plant 53:657–662
Takehisa H, Shimodate T, Fukuta Y, Ueda T, Yano M, Yamaya T, Kameya T, Sato T (2004) Identification of quantitative trait loci for plant growth of rice in paddy field flooded with salt water. Field Crops Res 89:85–95
Thomson MJ, Zhao K, Wright M, McNally KL, Rey J, Tung CW, Reynolds A, Scheffler B, Eizenga G, McClung A, Kim H, Ismail AM, Ocampo M, Mojica C, Reveche MY (2011) High-throughput single nucleotide polymorphism genotyping for breeding applications in rice using the BeadXpress platform. Mol Breed 29:875–886
Toda Y, Tanaka M, Ogawa D, Kurata K, Kurotani K, Habu Y, Ando T, Sugimoto K, Mitsuda N, Katoh E, Abe K, Miyao A, Hirochika H, Hattori T, Takeda S (2013) Rice salt sensitive 3 forms a ternary complex with JAZ and class-C bHLH factors and regulates Jasmonate-induced gene expression and root cell elongation. Plant Cell 25:1709–1725
Wu H, Ye H, Yao R, Zhang T, Xiong L (2015) OsJAZ9 acts as a transcriptional regulator in Jasmonate signaling and modulates salt stress tolerance in rice. Plant Sci 232:1–12
Yamaguchi T, Blumwald E (2005) Developing salt-tolerant crop plants: challenges and opportunities. Trends Plant Sci 10:615–620
Yang J, Sun K, Li D, Luo L, Liu Y, Huang M, Yang G, Liu H, Wang H, Chen Z, Guo T (2019) Identification of stable QTLs and candidate genes involved in anaerobic germination tolerance in rice via high-density genetic mapping and RNA-Seq. BMC Genomics 20:355
Zang J, Sun Y, Wang Y, Yang J, Li F, Zhou Y, Zhu L, Jessica R, Mohammadhosein F, Xu J, Li Z (2008) Dissection of genetic overlap of salt tolerance QTLs at the seedling and tillering stages using backcross introgression lines in rice. Sci China Ser C 51:583–591
Zhang Y, Lv Y, Jahan N, Chen G, Ren D, Guo L (2018) Sensing of abiotic stress and ionic stress responses in plants. Int J Mol Sci 19(11):3298
Acknowledgements
This study was supported by the National Science-technology Support Projects (Grant No. 2015BAD01B02-02), the Natural Science Foundation of China (Grant Nos. 31461143014, 31701390 and 31801336), the National Key Research and Development Program of China (Grant No. 2016YFD0100902-07).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jahan, N., Zhang, Y., Lv, Y. et al. QTL analysis for rice salinity tolerance and fine mapping of a candidate locus qSL7 for shoot length under salt stress. Plant Growth Regul 90, 307–319 (2020). https://doi.org/10.1007/s10725-019-00566-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-019-00566-3