Abstract
Panicle structure and grain shape are important components of rice architecture that directly contribute to rice yield and are regulated by quantitative trait loci (QTLs). In this study, a doubled haploid (DH) population derived from a cross between japonica “Maybelle” and indica “Baiyeqiu” was used to determine genetic effects on panicle structure and grain shape. All detected traits exhibited a continuous, transgressive distribution in the DH population. QTL analysis showed that a total of 24 QTLs related to panicle structure and grain shape were detected on chromosomes 1, 2, 3, 4, 5, 6, 7, 9, 10, and 12, and three epistatic interaction QTLs were detected. Some genes related to panicle structure and grain shape were predicted in the major QTLs, and variations existed between the parents in all genes. Only OsIDS1, GS5, and SRS3 had nonsynonymous mutations that led to protein changes. Quantitative real-time PCR analysis showed that the expression levels of GS5 and OsFOR1 significantly differed between the two parents. In addition, genetic diversity analysis showed that the H3-35–H3-37, H6-18–H6-19, and H7-12–H7-14 intervals might be selected in the breeding program.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
As an important staple food, rice plays a key role in feeding people worldwide. With the rapidly increasing global population and drastic decreases in cultivated land and water resources, improving rice productivity is a challenge for rice breeders and plant geneticists (Sakamoto and Matsuoka 2004). Producing rice with an ideal panicle structure is an important direction for super high-yield breeding, and 1000-grain weight which determined by grain shape can also directly affect rice yield (Wang et al. 2019). Therefore, both panicle structure and grain shape are important agronomic traits related to grain yield (Wu et al. 2016). Rice panicle structure usually refers to the size, number, and growth posture of rice panicles. Accordingly, both panicle branching and spikelet formation can influence the formation of panicle (Yu et al. 2017), and rice panicle structure estimation indices usually include the number of spikelets per panicle (SPP), panicle length (PL), and the numbers of primary branches (PBN) and secondary branches (SBN) (Sun et al. 2017). The broad concept of panicle structure includes grain shape, which can be inferred by grain length (GL), grain width (GW), and grain length–width ratio (LWR) (Bai et al. 2010).
Gene cloning and QTL analysis are beneficial for elucidating the genetic basis of rice yield-related traits. To date, lots of QTLs and genes related to panicle structure have been detected using an F2 population, a doubled haploid (DH) population, and a recombinant inbred line (RIL) population. Gna1, a QTL for number of grains per panicle, encodes cytokinin oxidase/dehydrogenase and regulates cytokinin degradation. Decreased expression of Gna1 led to cytokine accumulation in inflorescence meristems and increased the number of reproductive organs (Messenger et al. 2005). DEP1, a QTL for the number of rachis branches, is responsible for panicle type. The dominant allele at the DEP1 locus caused truncation of phosphatidylethanolamine-binding protein-like domain protein, which enhanced meristematic activity and resulted in reduced inflorescence internode length (Huang et al. 2009; Terao et al. 2010). IPA1, an ideal plant architecture gene, encodes SOUAMOSA PROMOTER BINDING PROTEIN-LIKE 14 (SPL14), which is regulated by microRNA OsmiR156 in vivo. Decreased expression of OsSPL14 caused a reduction in panicle branches, grain number, plant height, and culm diameter (Jiao et al. 2010). However, there were some genes cloned from mutants that regulate plant structure, such as APO1, LAX1, LAX2, FZP, and SP1 (Bai et al. 2016; Okada et al. 2018). APO1, which encodes the F-box protein, regulates the transformation of the meristem into the panicle primordial, and its overexpression caused an increase in the number of primary branches and spikelets, thereby enhancing rice yield (Ikeda et al. 2007). Both LAX1 and LAX2 are required for axillary meristem initiation and maintenance during panicle development (Oikawa and Kyozuka 2009; Tabuchi et al. 2011). FZP is a negative regulator of axillary meristem formation within the spikelet meristem and a positive regulator of floral meristem identity establishment (Komatsu et al. 2003). SP1 encodes a putative transporter of the peptide transporter family and influences branch elongation (Li et al. 2009). OsMADS34/PAP2, a SEPALLATA-like gene, encodes a MADS-box protein that contains a short carboxyl terminus without transcriptional activation activity. The mutant osmads34 displayed decreased spikelet number and modified spikelet morphology, which included altered lemma/leaf-like elongated sterile lemmas (Gao et al. 2010).
Several QTLs have been identified that are associated with grain length and grain weight, such as GS3 and GL3.1 (Fan et al. 2006; Qi et al. 2012), and grain width, such as GW2, GW5/GSE5 (Song et al. 2007; Weng et al. 2008; Liu et al. 2017). Truncation in the terminal of the predicted protein GS3 led to long rice grains. Recently, Xia et al. (2018) found that GL3.3 epistatically interacted with GS3 to affect rice grain length. GL3.1 encodes a phosphatase kelch family protein that controls protein phosphorylation in spikelets to accelerate cell division, which resulted in long grains (Qi et al. 2012). GW2 encodes a RING-type protein with E3 ubiquitin ligase activity. Thus, the loss of GW2 function produced more cells and larger spikelet hulls, which resulted in enhanced grain width. Weng et al. (2008) found that a 1212-bp deletion at the GW5 locus can affect grain width; a recent report revealed that this 1212-bp deletion is the promoter region of GSE5, and a 950-bp deletion was also identified in the same promoter region of some indica varieties. GSE5 encodes a plasma membrane-associated protein with the IQ domain, and this protein can interact with the rice calmodulin protein OsCaM1-1. Deletion at the promoter caused decreased GSE5 expression and resulted in wider grain width by influencing cell proliferation in spikelet hulls (Duan et al. 2017).
Although many QTLs and genes related to panicle structure and grain shape have been identified, it is necessary to identify more QTLs and genes to elucidate the molecular mechanisms of panicle structure and grain shape regulation, and to provide more information and genetic resources to improve rice plant architecture. In addition, such information can provide more insights into the history of yield-related traits development through natural evolution and human selection. Panicle structure and grain shape are critical for improving rice breeding programs. Selection may be the most important factor that has influenced panicle structure and grain shape in modern rice varieties. Furthermore, many related genes were also reported to be selected by humans in modern breeding (Wu et al. 2014; Win et al. 2017; Fan et al. 2006; Duan et al. 2017).
Here, we used a DH population derived from a cross between an improved variety, “Maybelle,” and a landrace variety, “Baiyeqiu,” to identify QTLs for panicle structure and grain shape. Genetic diversity analysis of these QTLs was also performed to study the effects of human selection on rice breeding programs. These results can elucidate the genetic basis and effects of rice breeding on panicle structure and grain shape.
Materials and methods
Plant material
A set of 168 DH lines developed by anther culture of an F1 hybrid between the typical local indica variety “Baiyeqiu” and the japonica cultivar “Maybelle” was used for QTL analysis (Yang et al. 2018). All lines were planted at the experimental farm of China National Rice Research Institute (Hangzhou, China). For field planting, one line was planted in each plot; four rows per plot and six individuals per row were planted. The natural population, which included 285 local varieties and 92 improved varieties, was used, and more information about this population was provided in a previous study (Xu et al. 2016).
Phenotypic evaluation
At harvest, we selected five plants in the middle of each line and picked the three highest panicles from each plant. Then, we measured PL, PBN, SBN, and SPP for the 15 panicles. After removing inordinate values from measuring data, we calculated the average of the values as the final phenotypic data of each trait. Following adequate drying, more than 100 fully filled grains were used to measure the GL and GW with an electronic digital display Vernier caliper.
RNA extraction and quantitative real-time PCR analysis
Total RNA was extracted from 2-cm young panicle tissues of the two parents “Baiyeqiu” and “Maybelle” using a Total RNA extraction kit (Takara, Japan). First-strand cDNA was synthesized using PrimerScript RT Master Mix (Takara, Japan). Then, quantitative real-time PCR (qRT-PCR) was performed in a two-step reaction using PowerUp SYBRGreen Master Mix (Applied Biosystems, USA) on an Applied Biosystems 7500 Real-Time PCR system (Applied Biosystems, USA). The mRNA expression was quantified for GS5 (LOC_Os05g06660), OsIDS1 (LOC_Os03g60430), OsFOR1 (LOC_Os07g38130), DST (LOC_Os03g57240), SRS3 (LOC_Os05g06280), and Ubq (LOC_Os03g13170). The rice ubiquitin gene was used as the internal control. Each measurement was performed on three replicates of each of three biological samples. The primers used for the qRT-PCR are listed in the supplemental materials (Table S1).
Data analysis
Statistical analyses of means, frequency statistics, tha values of skewness and kurtosis for the DH population, and a Student’s t-test between the parents were calculated using Microsoft Office Excel 2010 and SAS 9.2 (SAS, Inc., Cary, North Carolina, USA). Correlation analysis, including calculation of correlation coefficients between each pair of traits, was performed using the psych package in R 3.5.1 (https://personality-project.org/r/book/). Genetic diversity indices, which included both polymorphism information content (PIC) and reduction of diversity (ROD) for local and improved varieties, were calculated as described by a previous study (Yang et al. 2018). The genotype of every variety was also obtained by a previous report (Xu et al. 2016). The parents were sequenced using next-generation sequencing technology on the Illumina platform for about 20 × coverage of rice genome. All paired-end reads were aligned and mapped using BWA software (Li and Durbin 2009). SNP and INDEL calling were conducted using the GATK pipeline as previously described (McKenna et al. 2010).
Linkage map construction and QTL mapping
In total, 295 polymorphic markers between the parents, which included 187 INDEL markers and 108 SSR markers, were uniformly distributed on 12 rice chromosomes, and primers were designed using Primer Premier 3.0 (http://bioinfo.ut.ee/primer3-0.4.0/). Additionally, these polymorphic markers were used for genetic linkage map construction of the DH population with IciMapping (Meng et al. 2015).
Then, by combining the phenotypic data and linkage map, we used the inclusive composite interval mapping as our QTL mapping method to obtain the candidate QTLs (Wang 2009). The presence of a QTL was defined as the threshold of likelihood of odds (LOD) value greater than 2.5. In addition, the genetic parameter of additive effect and variance explained by each QTL were also calculated. The identified QTLs were named according to the standard nomenclature (McCouch et al. 1997).
Results
Phenotypic variation in the parents and DH lines
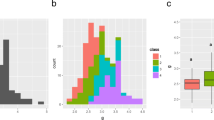
As the results shown in Fig. 1, all traits, which included PL, PBN, SBN, SPP, GL, GW, and LWR, exhibited a continuous distribution and substantial transgressive variations in the DH population. Except for SPP and GW, all other traits showed significant differences (p < 0.05) between the two parents. Moreover, “Maybelle” exhibited higher values than “Baiyeqiu” for all traits except PBN.
Skewness and kurtosis were calculated to describe the distribution of the traits in our research. As shown in Table 1, all traits had positive skewness; this might be because of the larger contribution of “Maybelle” alleles to the DH population. For kurtosis, all traits except for PL displayed a leptokurtic distribution (kurtosis value > 0).
Correlation analysis
Correlation analysis showed that there was no significant correlation between panicle structure traits and grain shape traits, because the correlation coefficients ranged from − 0.21 to 0.18, and this was especially true for correlations between PBN and grain shape traits, also between GW and panicle structure traits (Fig. 2). However, high correlations were observed between any two panicle structure traits and any two grain shape traits. Among these panicle structure traits, positive correlations were observed between any two traits. Additionally, the correlation coefficient between SBN and SPP was the greatest at 0.92. For grain shape traits, LWR was positively correlated with GL and negatively correlated with GW, and a high correlation were observed between LWR and GL with a correlation coefficient of 0.74.
QTL analysis
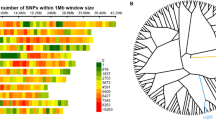
In total, 24 QTLs that contained 21 genetic intervals were detected for all traits (Table 2). These QTLs were distributed on chromosomes 1, 2, 3, 4, 5, 6, 7, 9, and 12 (Fig. 3). Fourteen QTLs were identified for panicle structure with LOD scores that ranged from 2.75 to 6.12; the amount of phenotypic variation explained ranged from 4.74 to 15.02%. Furthermore, 10 QTLs for grain shape were identified and had LOD scores that ranged from 2.73 to 15.64, and the amount of explained phenotypic variation ranged from 3.86 to 30.86%.
Among these QTLs for panicle structure, there were five QTLs for PL detected on chromosomes 2, 3, 4, and 7, with LOD scores that ranged from 2.75 to 6.13, and they explained 4.74–15.02% of the phenotypic variation. Four of the QTLs, qPL2, qPL3, qPL4-1, and qPL7, exhibited negative additive effects, and alleles from “Baiyeqiu” increased PL. qPL3 had the highest LOD score. qPL3 was located in the H3-7–H3-8 interval on chromosome 3 and accounted for 15.02% of the phenotypic variation; the allele from “Baiyeqiu” can increase PL by 1.72 cm. Only one QTL, qPBN12, was identified for PBN in the H12-19–H12-20 interval on chromosome 12; it explained 10.47% of the phenotypic variation, and the LOD score was 3.96. Based on the additive effects, we infer that the allele from “Maybelle” primarily contributed to the observed phenotype. For SBN, four QTLs, qSBN1, qSBN3, qSBN4, and qSBN7, located on chromosomes 1, 3, 4, and 7, respectively, explained 5.83–11.47% of the phenotypic variation and had LOD scores that ranged from 3.04 to 5.12. Three of the QTLs displayed negative additive effects, which indicate that the alleles from “Baiyeqiu” increased SBN. Similar results were observed in QTL analysis for SPP; four QTLs were mapped on chromosomes 1, 3, 4, and 7 (qSPP1, qSPP3, qSPP4, and qSBPP7, respectively), with LOD scores of 3.16–3.87 and explained variations of 6.75–8.09%. Moreover, most of these QTLs for SPP overlapped with or were near the intervals detected in QTL analysis for SBN. For example, qSBN1 was co-localized with qSPP1, and both qSBN7 and qSPP7 were located in the H7-12–H7-13 interval.
In grain shape analysis, two, two, and six QTLs were detected for LWR, GL and GW, respectively. qLWR7, the major QTL for LWR, was located in the H7-13–H7-14 interval on chromosome 7, displayed the highest LOD score (15.64), and explained 30.86% of the phenotypic variation. The allele from “Maybelle” increased LWR. Moreover, the H7-13–H7-14 interval was also responsible for GL (qGL7), which was also the most important QTL for this trait, had an LOD score of 11.57, and explained 25.19% of phenotypic variation. Positive additive effects indicated that the interval in “Maybelle” might increase GL by 0.46 mm. Furthermore, six QTLs for GW, qGW1, qGW3-1, qGW3-2, qGW6, qGW9, and qGW12, were identified on chromosomes 1, 3, 6, 9, and 12, respectively. These QTLs did not overlap with those for LWR, and explained 3.86–12.30% of the phenotypic variation with LOD scores that ranged from 2.73 to 6.60. Additionally, four of the QTLs (qGW1, qGW3-1, qGW3-2, and qGW6) displayed negative additive effects. qGW12 was the most important QTL for GW and was located in the H12-3–H12-4 interval on chromosome 12 with an LOD score of 6.60, and accounted for 12.30% of phenotypic variation.
Epistatic interaction analysis
To further understand the genetic components of all traits, the digenic epistatic effects were estimated. We detected epistatic interactions at three QTLs for only two traits (SBN and GL; Table 3). One pair of epistatic loci was detected for SBN, which explained 20.49% of the phenotypic variation and had the highest LOD score. Two pairs of epistasis loci for GL were identified and accounted for 16.64% and 9.31% of the phenotypic variation. However, no significant epitasis was detected for other traits, which indicates that the main effect QTL is the primary genetic component of these traits.
Genetic diversity analysis of major QTLs
Because one parent is a landrace variety and the other is an improved variety, differences between them may partly represent differences between landrace and improved varieties. Therefore, the major QTLs detected here were used to study the artificial selection of panicle structure and grain shape in rice breeding. A natural population that included 95 improved varieties and 285 landrace varieties was used to analyze genetic diversity differences between improved and landrace varieties. We observed some differences between improved and landrace varieties in the intervals identified in our study (Fig. 4a). The PIC values of the landrace varieties were somewhat higher than those of the improved varieties in certain intervals, including H3-35–H3-37, H6-18–H6-19, and H7-12–H7-14. Moreover, the ROD curve chart exhibited obvious peaks in these intervals (Fig. 4b). However, PIC and ROD analysis showed that there were no significant differences between landrace and improved varieties for other major QTLs detected. These findings indicate that the H3-35–H3-37, H6-18–H6-19, and H7-12–H7-14 intervals might be selected in the breeding program.
Genetic diversity analysis of the major QTL intervals in local and improved varieties. a Polymorphism information content (PIC) values of local and improved varieties in the major QTL regions. Blue and red curves show the PIC values of improved and local varieties, respectively. The black arrows indicate regions where the PIC value of local varieties was greater than that of improved varieties; b parameter of reduction of diversity (ROD) distribution at candidate QTL regions. Red dotted line means the boundary of two intervals.
Comparative genetic analysis for QTLs and trait-related cloned genes
Many genes related to panicle structure and grain shape were previously cloned in rice. By integrating the previously cloned genes and QTLs detected in this study, we further found that five related genes, OsIDS1, OsFOR1, DST, GS5, and SRS3, were located in the QTLs detected above (Fig. 5a). Among these genes, OsIDS1, OsFOR1, and DST were related to panicle structure, whereas GS5 and SRS3 were involved in grain shape regulation. Then, we compared the sequences of these genes between the two parents, and found that many differences (SNPs and INDELs) were found in all five genes, including in their promoter regions (Fig. 5a and Supplementary Table S2).
Diversity in OsIDS1, OsFOR1, DST, GS5, and SRS3 between “Maybelle” and “Baiyeqiu”. a Variations in OsIDS1, OsFOR1, DST, GS5, and SRS3 between the parents. Solid and empty boxes indicate exons and untranslated regions, respectively; the red lines indicate the differences between the two parents. b Expression levels of OsIDS1, OsFOR1, DST, GS5, and SRS3 in the parents. OsIDS1 (LOC_Os03g60430), OsFOR1 (LOC_Os07g38130), DST (LOC_Os03g57240), GS5 (LOC_Os05g06660), SRS3 (LOC_Os05g06280). Mean and SD values from qRT-PCR analysis were obtained with three biological replicates in one experiment. *p < 0.05; **p < 0.01 (Student’s t test).
Consequently, to determine whether these differences can influence gene function, the differences in the genes, including SNPs and INDELs, were annotated based on the Nipponbare reference genome (http://rice.plantbiology.msu.edu/index.shtml). The annotation results showed that variations between the parents were found in the exons all five genes (Table 4). However, most of the variations were synonymous mutations, and two genes (DST and OsFOR1) had no protein change between parents. However, we do not know whether some functional variations exist in the promoter and untranslated regions of these genes. Moreover, GS5 had a nonsynonymous mutation and amino acid change at the position of 98 bp, which resulted in a change from alanine to glycine between the two parents. OsIDS1 had a nonsynonymous mutation and amino acid change at the position of 1118 bp, with proline in “Maybelle” and leucine in “Baiyeqiu.” Moreover, SRS3 had the largest number of variations in its exon region. The nonsynonymous mutation at the position of 2420 bp resulted in a change from arginine to histidine. Additionally, “Baiyeqiu” had a four bases inserted at the position of 2479 bp in SRS3, which resulted in a frameshift mutation and premature termination.
Based on these results, qRT-PCR analysis was also performed on these related genes to compare the expression levels between the two parents (Fig. 5b). The results showed that the expression levels of OsFOR1 and GS5 significantly differed (p < 0.05) between the two parents, and the expression levels of DST, SRS3, and OsIDS1 showed no obvious differences. Moreover, “Baiyeqiu” exhibited higher expression of GS5 and OsFOR1 than “Maybelle”, which might affect the panicle structure and grain shape of the parents.
The results described above demonstrate that these five genes might contribute to panicle structure and grain shape, and be candidate genes for QTLs. However, further confirmation of these genes should be performed.
Discussion
The DH lines exhibited considerable variations for all traits evaluated in this study, and many QTLs related to these traits were also detected. The range and mean values of phenotypes reflected the extent of phenotypic variability in the DH population, which indicated that variations in panicle structure and grain shape were caused by multiple genes and their interactions. In the DH population, PBN displayed positive skewness, but “Maybelle” exhibited greater PBN than “Baiyeqiu”, this might because PBN is controlled by a few minor genes and is also affected by polygenes (Zhang et al. 2002; Jiang et al. 2010). Kurtosis of the phenotype distribution only displayed a negative value for PL, which indicates that PL distributed asunder. This finding may have resulted because the DH population has less chance for recombination of non-allelic genes or because PL is environmentally sensitive (Choo and Reinbergs 1982; Liao et al. 2000). Additionally, DH lines with phenotypic values greater than the parent with the higher value and less than the parent with the lower value were observed for all traits (transgressive segregation). This may indicate that different genes assembled and epistatic interactions led to different panicle structure and grain shape. Thus, these findings can provide information for ideal rice plant architecture planning and also benefit rice breeding by gene pyramiding to enhance grain yield.
Both panicle structure and grain shape were panicle-related traits, but they were relatively independent. Correlation analysis indicated that they were not correlated with each other and had low correlation coefficients. Furthermore, no common QTL was detected between these two types of traits. However, there was extremely significant correlation between any pairs of traits related to panicle structure or grain shape. Moreover, 25 QTLs that covered 21 intervals were identified in QTL analysis, which indicated that some intervals had two or more QTLs, and some relationships may exist among the clustered QTLs. Two intervals that contained four QTLs were responsible for SBN and SPP, and a common interval was detected for GL and LWR. QTLs in a single genetic interval that control multiple traits may help explain the high correlation coefficients between phenotypes of the pairs of traits. Similar findings were also reported in the context of bi-parental populations in a previous study (Mei et al. 2006) and may indicate that these intervals harbor QTLs that exhibit pleiotropism or physiological associations. As far as we know, increased SPP and SBN both enhanced rice yield; therefore, we might directly use these QTLs in marker-assisted selection for collaborative improvement of multiple traits.
Panicle structure and grain shape are very complicated characters in rice. Lots of QTLs for panicle structure have been identified in previous reports. Some of these QTLs were also detected in the same regions in this study, which might indicate that they represent the same functional loci. qSPP4 was located in the H4-9–H4-10 interval and overlapped with the previously reported QTL spp4-2, which is related to SPP and was identified from RILs derived from an indica/japonica cross between 9024 and LH422 (Xiao et al. 1996). Mei et al. (2006) reported a QTL, qSNP-4a, which regulated SPP in a reciprocal introgressive population; interestingly, it was found in the same region as qSBN4, which was located in the H4-5–H4-6 interval. Moreover, in the H1-7–H1-8 interval, the QTL for SBN and SPP was detected and overlapped with qSSB1-1, a QTL for SBN that was previously reported by Cui et al. (2002).
Some previously reported QTLs for grain shape were also detected in this study. For example, gw3.1, which was previously identified as a QTL for grain width using a backcross population with the parents Oryza rufipogon and O. sativa cultivar Jefferson, overlapped with the major QTL qGW3-2 (Thomson et al. 2003). qGW3, which was identified from a RILs cross between Koshihikari and IR24, was located in the H3-12–H3-13 interval (Miyata et al. 2007). Some cloned genes were also located in the QTLs we detected. OsIDS1, a gene cloned from the mutant, mapped in the H3-36–H3-37 interval and overlapped with qSBN3. It was reported that OsIDS1 plays an important role in inflorescence architecture and floral meristem establishment (Lee and An 2012). OsFOR1, which encodes a protein that contains a leucine-rich repeat domain, played a role in the formation and/or maintenance of floral organ primordia, and overlapped with the QTL qSPP7 (Jang et al. 2003). DST was found in the H3-35–H3-36 interval. DST can regulate OsCKX2/Gna1 expression and the phytohormone cytokinin level in the reproductive shoot apical meristem, and then consequently influence SPP (Terao et al. 2010). Gene sequence comparison revealed that the variations in these three genes between the parents could lead to protein alterations, and OsIDS1, OsFOR1, and DST were probably candidate genes in these three intervals. Considering the LOD scores of these QTLs, we found that OsIDS1 and DST were mapped in the major QTLs of SPB and SPP, respectively. Moreover, OsIDS1 had a nonsynonymous mutation between the two parent sequences. Thus, we speculated that OsIDS1 was probably a functional locus in the DH population; however, more research is still needed to verify this.
Importantly, all of the genes were cloned from mutants, and they were not identified in the natural varieties. This provided an opportunity to explore the function and evolution of these genes in rice breeding. The largest QTLs for grain shape were qLWR7 and qGL7, which were located in the same interval (H7-13–H7-14). Additionally, GL7, which encodes a protein homologous to Arabidopsis thaliana LONGIFOLIA proteins, was also mapped in this region (Wang et al. 2015). Therefore, we are confident that GL7 is a functional locus in the H7-13–H7-14 interval for two reasons. First, this interval had the major QTL for LWR and GL, which was consistent with the function of GL7. Second, it was previously reported that GL7 widely existed in the tropical japonica, which was geographically distributed across the America, and one of the parents in our study, “Maybelle,” was original planted in the America. Besides GL7, some loci detected overlapped with other previously reported genes. For example, GS5 (Xu et al. 2015) and SRS3 (Kitagawa et al. 2010) were anchored in the H5-4–H5-5 interval, which was responsible for LWR, and there was also substantial variation between in these two genes the parents. In addition, “Baiyeqiu” showed higher GS5 expression than “Maybelle” (Fig. 5b). The high expression of GS5 resulted in large rice grains, which was consistent with the findings of previous research (Xu et al. 2015). Moreover, by combining the gene diversity results between “Maybelle” and “Baiyeqiu,” we concluded that variations at the OsFOR1 and GS5 promoters may cause differences in expression between the two parents. The consistency of these results partly supports the robustness of our QTL analysis.
Some novel QTLs were also detected in our research, which may be related to different genetic backgrounds and environments. qPL3, qPL4-2, qGL6, and qGW12 displayed higher effective values than other novel QTLs, and explained approximately 10% of the phenotypic variation and had LOD scores greater than 4.5. These intervals should be fine-mapped by constructing a segregated population and marker density should be increased. Other novel QTLs had minor effects and no major QTL was detected nearby. Some novel epistatic QTLs were also detected and might work together to influence phenotype. Therefore, the effects of minor and epistatic QTLs should also be considered in the future.
In our study, “Maybelle” was an improved variety, whereas “Baiyeqiu” was a local variety. The different performances between these varieties might indicate that these traits were improved by selection. “Baiyeqiu” displayed larger panicles and grain size and enhanced rice production, which may be preferential for breeding. However, in modern rice breeding programs, the panicle and grain sizes similar to “Maybelle” are always selected because of the grain plumpness in giant rice grains and the balance between source and sink. Moreover, genetic diversity analysis showed that many QTLs detected in our study displayed higher genetic diversity in local varieties than in improved varieties, which indicated that some QTLs of panicle structure and grain shape from local varieties disappeared through human selection during rice breeding, and led to more suitable panicle structure and grain shape of improved varieties in rice breeding programs. These findings provided new insights into the genetic domestication mechanisms of rice panicle structure and grain shape and a theoretical basis for future rice breeding.
References
Bai XF, Luo LJ, Yan WH, Kovi MR, Zhan W, Xing YZ (2010) Genetic dissection of rice grain shape using a recombinant inbred line population derived from two contrasting parents and fine mapping a pleiotropic quantitative trait locus qGL7. BMC Genet 11:16
Bai XF, Huang Y, Mao DH, Wen M, Zhang L, Xing YZ (2016) Regulatory role of FZP in the determination of panicle branching and spikelet formation in rice. Sci Rep 6:19022
Choo TM, Reinbergs E (1982) Analyses of skewness and kurtosis for detecting gene interaction in a doubled Haploid population. Crop Sci 22:231–235
Cui KH, Peng SB, Xing Yz, Yu SB, Xu CG (2002) Genetic analysis of the panicle traits related to yield sink size of rice. Acta Agronomic Sinica 29:144–152
Duan PG, Xu JS, Zeng DL, Zhang BL, Geng MF, Zhang GZ, Huang K, Huang LJ, Xu R, Ge S et al (2017) Natural variation in the promoter of GSE5 contributes to grain size diversity in rice. Mol Plant 10:685–694
Fan CC, Xing YZ, Mao HL, Lu TT, Han B, Xu CG, Li XH, Zhang QF (2006) GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor Appl Genet 112:1164–1171
Gao XC, Liang WQ, Yin CS, Ji SM, Wang HM, Su X, Guo CC, Kong HZ, Xue HW, Zhang DB (2010) The SEPALLATA-like gene OsMADS34 is required for rice inflorescence and spikelet development. Plant Physiol 153:728–740
Huang XZ, Qian Q, Liu ZB, Sun HY, He SY, Luo D, Xia GM, Chu CC, Li JY, Fu XD (2009) Natural variation at the DEP1 locus enhances grain yield in rice. Nat Genet 41:494–497
Ikeda K, Ito M, Nagasawa N, Kyozuka J, Nagato Y (2007) Rice ABERRANT PANICLE ORGANIZATION 1, encoding an F-box protein, regulates meristem fate. Plant J 51:1030–1040
Jang S, Lee B, Kim C, Kim SJ, Yim J, Han JJ, Lee S, Kim SR, An G (2003) The OsFOR1 gene encodes a polygalacturonase-inhibiting protein (PGIP) that regulates floral organ number in rice. Plant Mol Biol 53:357–372
Jiang JH, Zhang QW, Hong DL (2010) Genetic analysis of panicle traits in Oryza sativa ssp. japonica. Chin Bull Bot 45:182–186
Jiao YQ, Wang YH, Xue DW, Wang J, Yan MX, Liu GF, Dong GJ, Zeng DL, Lu ZF, Zhu XD et al (2010) Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet 42:541–544
Kitagawa K, Kurinami S, Oki K, Abe Y, Ando T, Kono I, Yano M, Kitano H, Iwasaki Y (2010) A novel kinesin 13 protein regulating rice seed length. Plant Cell Physiol 51:1315–1329
Komatsu M, Chujo A, Nagato Y, Shimamoto K, Kyozuka J (2003) FRIZZY PANICLE is required to prevent the formation of axillary meristems and to establish floral meristem identity in rice spikelets. Development 130:3841–3850
Lee DY, An G (2012) Two AP2 family genes, supernumerary bract (SNB) and Osindeterminate spikelet 1 (OsIDS1), synergistically control inflorescence architecture and floral meristem establishment in rice. Plant J 69:445–461
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760
Li SB, Qian Q, Fu ZM, Zeng DL, Meng XB, Kyozuka J, Maekawa M, Zhu XD, Zhang J, Li JY, Wang YH (2009) Short panicle1 encodes a putative PTR family transporter and determines rice panicle size. Plant J 58:592–605
Liao CY, Wu P, Hu B, Ni JJ (2000) QTLs and epistasis underlying rice (Oryza sativa L.) panicle length in different genetic background and environments. Acta Genetics Sinica 27:599–607
Liu J, Chen J, Zheng X, Wu F, Lin Q, Heng Y, Tian P, Cheng Z, Yu X, Zhou K et al (2017) GW5 acts in the brassinosteroid signalling pathway to regulate grain width and weight in rice. Nat Plants 3:17043
McCouch SR, Cho YG, Yano M, Paul E, Blinstrub M, Morishima H, Kinoshita T (1997) Report on QTL nomenclature. Rice Genet Newsl 14:11–131
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M et al (2010) The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303
Mei HW, Xu JL, Li ZK, Yu XQ, Guo LB, Wang YP, Ying CS, Luo LJ (2006) QTLs influencing panicle size detected in two reciprocal introgressive line (IL) populations in rice (Oryza sativa L.). Theor Appl Genet 112:648–656
Meng L, Li HH, Zhang LY, Wang jK (2015) QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. The Crop Journal 3:269–283
Messenger S, Keller LP, Lauretta DS (2005) Cytokinin Oxidase Regulates Rice Grain Production. Science 309:737–741
Miyata M, Yamamoto T, Komori T, Nitta N (2007) Marker-assisted selection and evaluation of the QTL for stigma exsertion under japonica rice genetic background. Theor Appl Genet 114:539–548
Oikawa T, Kyozuka J (2009) Two-step regulation of LAX PANICLE1 protein accumulation in axillary meristem formation in rice. Plant Cell 21:1095–1108
Okada S, Sasaki M, Yamasaki M (2018) A novel rice QTL qOPW11 associated with panicle weight affects panicle and plant architecture. Rice (N Y) 11:53
Qi P, Lin YS, Song XJ, Shen JB, Huang W, Shan JX, Zhu MZ, Jiang L, Gao JP, Lin HX (2012) The novel quantitative trait locus GL3.1 controls rice grain size and yield by regulating Cyclin-T1;3. Cell Res 22:1666–1680
Sakamoto T, Matsuoka M (2004) Generating high-yielding varieties by genetic manipulation of plant architecture. Curr Opin Biotechnol 15:144–147
Song XJ, Huang W, Shi M, Zhu MZ, Lin HX (2007) A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat Genet 39:623–630
Sun B, Zhan XD, Lin ZC, Wu WX, Yu P, Zhang YX, Sun LP, Cao LY, Cheng SH (2017) Fine mapping and candidate gene analysis of qHD5, a novel major QTL with pleiotropism for yield-related traits in rice (Oryza sativa L.). Theor Appl Genet 130:247–258
Tabuchi H, Zhang Y, Hattori S, Omae M, Shimizu-Sato S, Oikawa T, Qian Q, Nishimura M, Kitano H, Xie H et al (2011) LAX PANICLE2 of rice encodes a novel nuclear protein and regulates the formation of axillary meristems. Plant Cell 23:3276–3287
Terao T, Nagata K, Morino K, Hirose T (2010) A gene controlling the number of primary rachis branches also controls the vascular bundle formation and hence is responsible to increase the harvest index and grain yield in rice. Theor Appl Genet 120:875–893
Thomson MJ, Tai TH, McClung AM, Lai XH, Hinga ME, Lobos KB, Xu Y, Martinez CP, McCouch SR (2003) Mapping quantitative trait loci for yield, yield components and morphological traits in an advanced backcross population between Oryza rufipogon and the Oryza sativa cultivar Jefferson. Theor Appl Genet 107:479–493
Wang JK (2009) Inclusive composite interval mapping of quantitative trait genes. Acta Agronomic Sinica 35:239–245
Wang Y, Xiong G, Hu J, Jiang L, Yu H, Xu J, Fang Y, Zeng L, Xu E, Xu J et al (2015) Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat Genet 47:944–948
Wang W, Xu X, Zhu CW, Gu JF, Zhang WY, Liu G, Zhu JG (2019) Elevated CO2-induced changes in cytokinin and nitrogen metabolism are associated with different responses in the panicle architecture of two contrasting rice genotypes. Plant Growth Regul. https://doi.org/10.1007/s10725-019-00511-4
Weng J, Gu S, Wan X, Gao H, Guo T, Su N, Lei C, Zhang X, Cheng Z, Guo X et al (2008) Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res 18:1199–1209
Win KT, Yamagata Y, Doi K, Uyama K, Nagai Y, Toda Y, Kani T, Ashikari M, Yasui H, Yoshimura A (2017) A single base change explains the independent origin of and selection for the nonshattering gene in African rice domestication. New Phytol 213:1925–1935
Wu T, Shen Y, Zheng M, Yang C, Chen Y, Feng Z, Liu X, Liu S, Chen Z, Lei C et al (2014) Gene SGL, encoding a kinesin-like protein with transactivation activity, is involved in grain length and plant height in rice. Plant Cell Rep 33:235–244
Wu Y, Fu Y, Zhao S, Gu P, Zhu Z, Sun C, Tan L (2016) CLUSTERED PRIMARY BRANCH 1, a new allele of DWARF11, controls panicle architecture and seed size in rice. Plant Biotechnol J 14:377–386
Xia D, Zhou H, Liu R, Dan W, Li P, Wu B, Chen J, Wang L, Gao G, Zhang Q, He Y (2018) GL3.3, a novel QTL encoding a GSK3/SHAGGY-like kinase, epistatically interacts with GS3 to produce extra-long grains in rice. Mol Plant 11:754–756
Xiao J, Li J, Yuan L, Tanksley SD (1996) Identification of QTLs affecting traits of agronomic importance in a recombinant inbred population derived from a subspecific rice cross. Theor Appl Genet 92:230–244
Xu CJ, Liu Y, Li YB, Xu XD, Xu CG, Li XH, Xiao JH, Zhang QF (2015) Differential expression of GS5 regulates grain size in rice. J Exp Bot 66:2611–2623
Xu Q, Yuan XP, Wang S, Feng Y, Yu HY, Wang YP, Yang YL, Wei XH, Li XM (2016) The genetic diversity and structure of indica rice in China as detected by single nucleotide polymorphism analysis. BMC Genet 17:53
Yang YL, Zhang MC, Xu Q, Feng Y, Yuan XP, Yu HY, Wang YP, Wei XH (2018) Exploration of genetic selection in rice leaf length and width. Botany 96:249–256
Yu HP, Qiu ZN, Xu QK, Wang ZW, Zeng DL, Zhang HJ, Gao GH,ZL, Chen ZY Guang et al (2017) Fine mapping of LOW TILLER 1, a gene controlling tillering and panicle branching in rice. Plant Growth Regul 83:93–104
Zhang ZH, Chen MM, Tang j, Hu ZL (2002) Genetic analysis of neck-panicle vascular bundle and panicle characteristics in rice (Oryza sativa L.). Acta Agronomic Sin 28:86–89
Acknowledgements
The present investigation is financially supported by National Key Research and Development Program of China (2016YFD0100101-02) and National Natural Science Foundation of China (31600999) and Chinese Academy of Agricultural Sciences (CAAS-ASTIP-201X-CNRRI).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Xu, X., Zhang, M., Xu, Q. et al. Quantitative trait loci identification and genetic diversity analysis of panicle structure and grain shape in rice. Plant Growth Regul 90, 89–100 (2020). https://doi.org/10.1007/s10725-019-00549-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-019-00549-4