Abstract
Genetic diversity analysis is pivotal for optimizing the use of genetic resources and deploying conservation strategies in the face of global changes. This is important for essential indigenous species often neglected by research. This research was carried out to examine the pattern of genetic diversity and structure among fonio (Digitaria exilis (Kippist) Stapf) accessions in Ghana. Following farmer surveys and collection efforts in key fonio-growing regions of Ghana, we amassed a comprehensive collection of 176 fonio accessions from 165 farmers across 24 communities. Farmer surveys identified 21 distinct cultivars cultivated predominantly for home consumption, averaging six cultivars per ethnic group. Out of the 176 accessions, 140 underwent genotyping using 14 simple sequence repeat (SSR) nuclear markers which revealed greater diversity within cultivars (58.5%) compared to between cultivars (16.8%) following molecular variance analysis. The organizational pattern of fonio genetic diversity was significantly influenced by farmer-specific cultivars, emphasizing the role farmers play in long-term preservation of fonio genetic diversity. Future strategies for the management, conservation, and utilization of fonio genetic resources in Ghana should incorporate traditional farming practices. Our findings further revealed the uniqueness of Ghanaian fonio genetic diversity, despite a smaller cultivation area compared to the global diversity. This shows the contribution of the interaction between the crop, environment and societal factors to the distinct genetic makeup observed in the Ghanaian fonio population. The observed genetic diversity can be exploited to identify and select genetically distinct individuals to develop tailored product profiles within fonio breeding programs for diverse market segments.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Genetic diversity analysis is an important pre-breeding activity geared towards deployment of efficient conservation strategies and effective utilization of genetic resources in a changing world (Adjebeng-Danquah et al. 2020; Govindaraj et al. 2015). This is particularly important for indigenous species which have been neglected by research although they have the potential to play numerous roles regarding global change.
Fonio (Digitaria exilis (Kippist) Stapf) stands out among neglected crops as a vital grain cereal, indigenous to West Africa, where millions rely on it for essential food and nutrition security (Adoukonou-Sagbadja et al. 2006). This crop plays a pivotal role in the cropping and food systems of numerous West African farm families. Its significance becomes evident in mitigating hunger during food shortages, as most fonio cultivars exhibit early maturation, ensuring a timely food source for farmers while waiting for the harvest of other staple crops (Vodouhe et al. 2007; Adoukonou-Sagbadja et al. 2006; Diop et al. 2018). Fonio’s importance transcends its role as a staple; it is recognized as a climate-smart crop, demonstrating resilience in the face of intermittent drought and thriving in environments with marginal soils and poor fertility (Vodouhe et al. 2007).
While fonio is acknowledged for its potential to be a crucial asset for smallholder farmers in adapting to global changes, the genetic diversity of this crop remains inadequately understood, especially in Ghana, where comprehensive research are notably lacking. Within the country, fonio production is predominantly concentrated in the northeastern region, where it holds significance as a favored crop among ethnic groups such as Anufor, Konkomba, Kabre, Dagomba, and Bassare. (Clottey et al. 2006a; Addai et al. 2022). Fonio is grown for both subsistence and commercial purposes. Generally, fonio production relies on traditional methods such as seed broadcasting and with low external input use. For many farmers, then, fonio is cultivated mainly for home consumption even though surplus grains are sold for income. In Ghana, commercial fonio production is experiencing a steady rise, driven by the active involvement of companies engaged in the comprehensive cycle of production, processing, and marketing of fonio. These enterprises are strategically positioned to meet both local and international demands for this valuable cereal. The international demand is increasing notably due to the awareness of the high nutrient composition and the numerous uses of the grains (Jideani and Jideani 2011).
The increasing local and international demands for fonio grains call for new market-oriented breeding programs. But for now, those programs cannot be implemented due to a lack of biological knowledge on fonio and on its genetic diversity. Indeed, there is little information on the genetic diversity of the Ghanaian fonio accessions and no fonio cultivar has been released and or registered for cultivation in Ghana. An earlier investigation to understand the Ghanaian fonio genetic diversity focussed on 13 accessions only, using agro-morphological markers (Clottey et al. 2006b). Later some accessions from Ghana were either resequenced (Abrouk et al. 2020) or genotyped with simple sequence repeat (SSR) markers for large scale characterization of fonio genetic diversity and population structure across West Africa (Kaczmarek et al. 2023). Kaczmarek et al. (2023) analyzed the genetic diversity of 1,539 accessions genotyped, of which 118 originated from Ghana. Interestingly, this analysis revealed a distinctiveness of some of these Ghanaian accessions as compared to other producing countries. But to date, no research has specifically focused and analyzed the genetic diversity and associated knowledge of fonio in Ghana. This calls for an in-depth characterization of Ghanaian fonio.
The aim of this paper is to describe the genetic diversity of Ghanaian fonio. It proposes to explore usual genetic diversity parameters such as the number of alleles, the allele frequencies, the observed (Ho) and expected heterozygosity (He). It pays specific attention to the organization of the fonio genetic diversity, considering biological, environmental and social factors. Recently, Diop et al. (2023) noted in Senegal that the spatial organization of genetic diversity of fonio was associated with farmers’ linguistic family. This is also the case for other crops where recent investigations have highlighted the influence of socio-cultural factors such as ethnicity and farmers’ seed exchange systems (Leclerc and Coppens d'Eeckenbrugge 2011; Delêtre et al. 2011; Labeyrie et al. 2014; Naino Jika et al. 2017; Orozco-Ramírez et al. 2016).
The local Ghanaian context of fonio cultivation raises many questions. Is fonio genetic diversity structured as a function of ethnic groups? Is it resulting from isolation by distance? Is it partitioned according to farmers’ cultivars? This last question is crucial as it is associated with the dynamics of in-situ fonio conservation in Ghana. A better understanding of in-situ fonio diversity at the local scale considering social factors is crucial to understand the evolutionary social process shaping fonio genetic diversity and thus to define conservation and breeding strategies.
Materials and methods
Farmer survey and germplasm collection
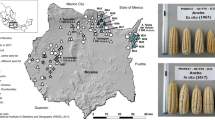
Farmer survey and fonio germplasm collection were conducted in the main fonio-growing areas of Ghana, in the North East region in June 2021 (Fig. 1, one district, Chereponi) and the Northern region in December 2021 (Fig. 1, four districts, Gushegu, Saboba, Tatale and Zabzugu), in collaboration with staff of the Department of Agriculture, who served as interpreters and helped us to define the sampling strategy. The purposive sampling strategy, a non-probability sampling method where units are selected because they possess characteristics that are needed in the sample, was used to target fonio growing communities in the five districts. Within these five districts, 24 communities actively engaged in fonio cultivation were targeted. Only farmers who cultivated fonio in the previous season, and thus considered to be active growers and could provide seed samples were interviewed.
Map of Africa highlighting fonio producing countries (A), map of Ghana with fonio producing regions (B, North East region in yellow; Northern region in orange), district map of Ghana showing the locations of the fonio germplasm collection (C) fonio production fields in the Chereponi district (D, E, ©Anthony Kojo, Chereponi, Ghana)
Community entry sensitization was carried out with the community leaders to seek their consent before conducting the surveys in the villages. The same approach was performed for each farmer providing information and/or seed samples. The farmers signed an informed consent form for each accession collected enabling their use for research purposes. Personal information and identity of the respondents were anonymized in the database.
A total of 165 farmers were surveyed across 24 communities, in five districts in the area where fonio is a popular crop (Chereponi, Saboba, Zabzugu, Gushegu, and Tatale). Those farmers belong to four ethnic groups, namely Anufor (121 farmers mainly from Chereponi district), Konkomba (30 farmers mainly from Saboba district), Kabre (10 farmers from Zabzugu district), and Dagomba (4 farmers from Gushegu district).
Farms were located at similar elevation above sea level (152 ± 50 m at 0.95 CI). Farmers surveyed represent all the age classes, from 20 to 60, with 15 farmers over 60 years. Mainly men were interviewed (142 men against 34 women). Men oversaw fonio selection, field preparation, seed sowing, harvesting, and threshing while women oversaw dehusking, winnowing and cooking (Pers. Obs., Richard Yaw Agyare). A semi-structured questionnaire was used to obtain socio-cultural information such as ethnicity, age, gender, marital status, educational level, etc. Their geographical location was recorded using a hand-held GPS device (Garmin GPSMAP 64X). In addition, knowledge on fonio cultivation, e.g. local cultivar names, phenology, and noteworthy traits and years of cultivation were captured. Origin of the first seed lots for each farmers’ cultivar, and source of seed used in the last season were also noted.
Fonio seeds were sampled from the seed stock of the farmers (harvested from the previous season) preserved into zip-lock plastic bags and labeled. A total of 176 accessions were collected from 165 farmers in 24 communities, constituting the largest fonio germplasm collection in Ghana (Table 1). The accessions were first planted in an observational trial and were thoroughly observed to eliminate off-type plants. Single plant selections were then made per accession from which seeds were harvested and multiplied for other research purposes.
The fonio accessions are conserved at CSIR-Savanna Agricultural Research Institute and duplicates were added to the global collection conserved at the Biological Resource Center, Seeds Adapted to Mediterranean and Tropical conditions, CIRAD-INRAE, ARCAD, Montpellier-France, for long-term conservation.
Sample preparation, DNA extraction and SSR genotyping
The 176 fonio accessions were grown in a growth chamber. Fresh leaf samples from fully opened leaves were sampled from a single plant per accession. Total genomic DNA extractions were performed from dried leaves by an automated method adapted from Risterucci et al. (2009) on Biomek FXP (Beckman Coulter, CA, USA) and using the NucleoMag Plant Kit (Macherey–Nagel, Germany).
DNA extraction and genotyping were carried out at the AGAP Institut laboratory, CIRAD, Montpellier, France. A total of 14 simple sequence repeat nuclear markers (Barnaud et al. 2012) were used for the genotyping (Table S1). PCR amplification was carried out in an ABI PRISM 3730 DNA Analyzer (Applied Biosystems, Foster City, CA, USA) while the DNA bands were sized with the GeneScan 600 LIZ Size Standard (Applied Biosystems, Foster City, CA). The PCR products were manually scored and verified twice with GeneMapper version 6 (Applied Biosystems, Foster City, CA, USA). Fonio accessions presenting more than three missing SSR genotypes were removed from the genetic analyses. Genetic analysis was thus based on 140 accessions, whereas farmer and social data analysis was based on 176 accessions.
Data analysis
Data were analyzed using packages in R software version 4.2.2 (R Core Team 2022).
Genetic diversity analysis
To estimate the level of genetic diversity of each SSR locus among the fonio accessions, the adegenet v. 2.1.10 (Jombart 2008) and pegas v. 1.2 (Paradis 2010) R packages were used to compute the number of alleles, the allele frequencies, the observed (Ho) and expected heterozygosity (He). Polymorphism information content was computed according to Botstein et al. (1980). The number of alleles, observed and expected heterozygosity and allelic richness (Ar) were again computed according to group level—community and farmers’ ethnic group and farmers' cultivar—with the matrixStats v. 0.63.0 (Bengtsson et al. 2023) R package. To account for the variability in group sizes, allelic richness was computed by averaging the calculation of 1000 randomly drawn iterations of four accessions and was not calculated for groups with less than four accessions. Allelic richness was compared using pairwise t-tests (1000 iterations, with Bonferroni p adjusted method). The mean inbreeding coefficient, FIS, was estimated with hierfstat v. 0.5-11 (Goudet and Jombart 2022) R package and the significant deviation from 0 was assessed using 1000 iterations over loci with the boot.ppfis function. We identified with a home-made script (Script R1) group of accessions having the same genotype at each locus, i.e. duplicate accessions.
Genetic structure analysis
Principal component analysis (PCA) was performed with the dudi.pca function in the ade4 v. 1.7-22 (Dray and Dufour 2007) R package. Data were first scaled with the scaleGen function in the adegenet v. 2.1.10 (Jombart 2008) R package and missing data were replaced with mean allele frequencies. The ggplot2 v. 3.4.2 (Wickham 2016) and factoextra v. 1.0.7 (Kassambara and Mundt 2020) R packages were used to plot the PCA.
We used the sparse Non-negative Matrix Factorization (sNMF) method implemented in the LEA v. 3.10.2 (Frichot and Francois 2015) R package for population structure analysis. We ran 10 independent repetitions for each K value (1–10) with a maximal number of iterations of 200. The cross-entropy criterion (lowest cross-entropy) was used to determine the optimal number of ancestral populations (Fig S1) and membership of groups was based on 0.70 or higher ancestry coefficient. Individuals with less than 0.70 ancestry coefficient for a group were considered as admixed. To validate the population structure among the fonio accessions, we conducted a discriminant analysis of principal components with the dapc function implemented in adegenet v. 2.1.10 (Jombart et al. 2010) R package. Structuring was done without a priori grouping (K-means clustering) of the accessions with defined factors. The find.clusters function in adegenet v. 2.1.10 (Jombart et al. 2010) R package was used to run successive K-means with 100,000 iterations for each run. We retained 50 PCs (chosen interactively from the console) based on the graph of cumulative variance explained by the eigenvalues of the PCA at the plateau level (Fig S2A). Also, four linear discriminants were retained based on the discriminant analysis eigenvalues. The measure of goodness of fit, Bayesian Information Criterion (lowest BIC), computed for each model was used to select the optimal number of K clusters that best described the population (Fig S2B). We compared the structuring based on DAPC and sNMF methods using a contingency table.
We conducted Chi2 tests with Monte-Carlo simulations to detect significant associations between genetic clusters inferred with sNMF and social or biological factors. We also performed Fisher’s exact test which produced a similar output as the Chi2 test with Monte-Carlo simulations. We performed an analysis of molecular variance with the poppr v 2.9.4 (Kamvar et al. 2014) R package to analyse how much of the variance is explained by the different social components of the fonio accessions (ethnicity of the farmer, farmers’ cultivar). Finally, to avoid confounding factors, we focused on the Anufor ethnic group (100 accessions) and conducted a hierarchical cluster analysis according to neighbor joining clustering of Nei’s genetic distance with the poppr v. 2.9.4 (Kamvar et al. 2014) R package. We then used a tree representation using the ggtree v. 3.6.2 (Yu 2022) R package.
Isolation by distance
The extent of fonio seed exchange was assessed by testing for isolation-by-distance (IBD) patterns, that is, we assessed whether genetic differentiation (FST)/(1- FST) among communities was correlated with geographical distance (Rousset 1997). We estimated the genetic distance FST using the hierfstat v. 0.5-11 (Goudet and Jombart 2022) R package. The genetic distance matrix (FST)/(1- FST) was plotted against the geographical distance using MASS v. 7.3-58.1 (Venables and Ripley 2010) and geodist v. 0.0.8 (Padgham 2021) R packages. A total of 1,000 Mantel tests were conducted with the adegenet v. 2.1.10 (Jombart 2008) R package between the two distant matrices to determine whether the relationship between genetic and geographical distances between communities was at random or not.
Comparison of the Ghanaian fonio germplasm with the global fonio diversity
To compare the genetic diversity in Ghana to the global fonio diversity, we retrieved the genetic dataset produced by Kaczmarek et al. (2023) consisting of 1539 fonio accession. We first selected the 118 Ghanaian accessions common in the two investigations. To have a balanced sampling with Ghana and a representative fonio cultivation area, two other sets of 118 accessions were selected with the kenstone algorithm from prospectr v. 0.2.6 (Stevens and Ramirez-Lopez 2022) R package that allows the sampling of a subset of accessions by maximizing the geographic range of their distribution. We ran the algorithm on the Western part (Senegal, Mali, Burkina Faso, Ivory Coast, Guinea) and Eastern part (Togo, Benin, Niger, Nigeria) of fonio geographic distribution. We then performed a PCA analysis on a total of 354 accessions originating from nine countries.
Results
This research aims to describe and analyze the genetic diversity of Ghanaian fonio at the local scale. It pays attention to how farmer-held cultivars contribute to maintaining fonio genetic diversity over time. The first section summarizes the main characteristics of farmers’ cultivars of fonio that were cultivated in Ghana. The second section presents the genetic parameters that characterize the fonio diversity. The third section pays attention to the organization of the fonio genetic diversity, considering geographical, biological and social factors, with more details about how farmers’ cultivars contribute to maintaining local fonio genetic diversity.
Characteristics of the farmers’ cultivars of fonio
In the research area, 21 farmers’ cultivar of fonio were inventoried (Fig S3). Majority of the 165 farmers surveyed (94% of farmers) cultivates one cultivar, with a mean of six cultivars per ethnic group. Fonio is mainly grown by men (81%) irrespective of the age class (Table S2). Cultivar richness is higher among Anufor farmers (nine cultivars), as compared to Konkomba (seven cultivars), Kabre (six cultivars) and Dagomba (one cultivar). Each farmers’ cultivar belongs almost exclusively to one ethnic group. Anufor and Konkomba farmers have been cultivating fonio for a longer period (average of 20 and 15 years, respectively) than Kabre and Dagomba farmers (average of 3 and 1 years, respectively). About 45% of the fonio cultivars are considered by farmers to have high yield, 24% to be easy to dehusk. The nutritional value of fonio as a noteworthy trait in a cultivar was not mentioned, except by a few farmers. The length of the cultivation cycle is 3 months for 56% of cultivars; 18% are less and 26% over 3 months.
Names of farmers’ cultivars are usually motivated (Table S3) with an explicit reference to ethnic groups (Namba and Ukabreja refer to the Namba and Ukabreja ethnic groups in Togo), to people (Yadema refers to the name of the former president of Togo, Gnassingbe Eyadema), or to notable morphological features. For instance, among the Konkomba ethnic group, some of the names are based on their distinctive pericarp color such as Ipui bolne (black skin fonio), Ipui maln (red skin fonio) and Ipui piln (white skin fonio). Names such as Fefeke and Kafia or Semiri connotes early maturity in Anufor and Kabre respectively.
Fonio is cultivated mainly for household consumption. 59.5% of the collected seed lots were produced for food while 36.9% were produced for both food and income, and 3.6% solely for sale (Table S4). Source of seeds for planting was mainly obtained from the household. Indeed, 89.7% of seed lots were self produced by farmers, while 7.3% were obtained from a company, who had previously obtained them from the Chereponi district, and 3% from friends in neighboring communities (Table S4).
By tracing for each cultivar, the origin of the initial seed lots, a diverse range of sources have been recorded. The origin of the initial cultivar seed lots corresponds to the sources through which the cultivars that are cultivated today were initially acquired. Among the Anufor, about 56% of farmers indicated that their initial cultivar seed lots originated from Togo, and 2% originated from Côte d’Ivoire. 42% of farmers do not know from which source they initially obtained the cultivar’s seed lots (Table S4). For Dagomba farmers, the origin of initial seed lots was traced back to a single company involved in the production and marketing of fonio in Ghana. The majority of Konkomba farmers (85.2%) do not know from which source they obtained their first seed lots. 11.1% were obtained from a company, and 3.7%, representing only one seed lot, from Togo. Similarly, among Kabre farmers, most of their cultivar seed lots were initially obtained from a company, 11.2% from Togo, and 33.3% of seed lot origin were unknown.
Genetic diversity of fonio
Out of the 14 SSR markers, two were monomorphic and were not considered further in the analyses. A total of 79 alleles were observed with an average of 3.27% missing data per locus (Table 2). The number of alleles per locus ranged from 2 to 24 with a mean of 6.58 alleles per locus.
The cumulative number of alleles per sample reached a plateau from 130 samples, suggesting that the sampling effort was sufficient (Fig S4). We identified 33 duplicates partitioned in nine groups, i.e. accessions genetically identical according to the marker used, among the accessions (Table S6). It ranged from two to ten accessions per group, and six groups gathered accessions originated from at least two districts.
SSR markers De_15, De_17 and De_38 exhibited the lowest number of alleles per locus (2) while locus De_04 showed the highest (24). Major allele frequency ranged from 0.22 (De_37) to 0.99 (locus De_15). Observed heterozygosity (Ho) varied from 0 to 0.22 with a mean of 0.08 and expected heterozygosity (He) from 0.03 to 0.89 with a mean of 0.37. SSR locus De_24 (0.22) had the highest observed heterozygosity while the highest expected heterozygosity was observed from locus De_37 (0.89). Polymorphism information content (PIC) ranged from 0.03 (De_15, two alleles with a MAF of 0.99) to 0.88 (De_37, 19 alleles with a MAF of 0.22) with a mean of 0.35. Locus De_37 (0.88) was the most polymorphic, followed by De_04 (0.68).
At the community level, the highest average number of alleles per locus was observed in Ando Nyamanu (3.83 alleles) while the lowest was observed in Sanguli (one allele) (Table 3). Significant differences (t-test, adj. p < 0.05) in observed heterozygosity existed between Gbangbanpong (0.23) and all the communities except Nayugu and Wonjuga with observed heterozygosity of 0.13 and 0.18 respectively. Expected heterozygosity ranged from zero (Sanguli) to 0.43 (Toma). Significant difference (t-test, adj. p = 0.0278) in expected heterozygosity existed between only Toma and Sanguli. Allelic richness was computed for communities with at least 4 accessions and ranged between 1.46 (Banjani) and 2.34 (Toma). Significant differences (t-test, adj. p < 0.001) in allelic richness existed among the communities except for Ando Nderenu versus Kudani, Ando Nyamanu versus Liwalbu, Bulasu versus Liwalbu, Ando Nderenu versus Naturi, Kudani versus Naturi, Kpalgabini versus Sangbana, Gbangbanpong versus Tosala Kombonli, Kpalgagbeni versus Wonjuga and Sangbana versus Wonjuga (t-test, adj. p > 0.05). In the case of FIS, the lowest average FIS was observed in Gbangbanpong (0.37, confidence interval = 0.95).
At the ethnicity level, the lowest average number of alleles per locus was recorded from farmers belonging to the Dagomba ethnic group (2.08) while the highest was from farmers belonging to Anufor ethnic group (5.75). The average number of alleles per locus among accessions from Konkomba and Kabre ethnic groups were 3.92 and 2.50 respectively. Observed heterozygosity ranged from 0.06 (Konkomba) to 0.09 (Anufor and Kabre). Expected heterozygosity was highest among accessions collected from the Konkomba ethnic group (0.45). However, no significant differences were observed among accessions from the ethnic groups for both observed and expected heterozygosity. Allelic richness significantly differed (t-test, adj. p < 0.001) among the ethnic groups except for accessions obtained from Dagomba and Konkomba farmers (t-test, adj. p > 0.05). Allelic richness ranged from 1.83 to 2.10 for Anufor and Konkomba ethnic groups respectively. In the case of inbreeding coefficient, higher values were observed within each ethnic group.
At the farmers’ cultivar level, the average number of alleles per locus ranged from 1.00 (Ipui Ukabreja, Kpentike, Yahran and No idea) to 5.42 (Yadema). No significant differences (t-test, adj. p > 0.05) were observed among the farmers’ cultivars for observed heterozygosity (Table 3). It ranged from 0.00 (Ipui Ukabreja, Yahran and No idea) to 0.27 (Fieja and Senbre). In the case of expected heterozygosity, it varied from 0.00 to 0.45. Significant differences (t-test, adj. p < 0.05) in expected heterozygosity were observed among Ipui maln and Ipui Ukabreja (t-test, adj. p < 0.01), Ipui maln and Kafia (t-test, adj. p < 0.04), Ipui maln and No idea (t-test, adj. p < 0.01), Ipui maln and Yahran (t-test, adj. p < 0.01), Ukabreja and Ipui Ukabreja (t-test, adj. p < 0.01), Ukabreja and No idea (t-test, adj. p < 0.01) and Ukabreja and Yahran (t-test, adj. p < 0.01). Allelic richness significantly differed among the farmers’ cultivars except between Ipui piln (1.83) and Namba (1.86). It ranged from 1.13 (Ukabreja) to 2.32 (Ipui maln). The lowest inbreeding coefficient was observed in Ipui (0.52) while the highest (0.94) was observed in Ipui piln.
Genetic structure analysis
Principal component analysis
Principal component analysis (PCA) revealed that 13.8% of the total variability among the fonio accessions was accounted for by the first two dimensions (Fig. 2). The first dimension of the plot (8.9%) differentiated the accessions into two groups. The first group was dominated by accessions collected from farmers belonging to the Anufor ethnic group (Fig. 2a). Majority of these accessions mainly corresponded to the Yadema farmers’ cultivar (Fig. 2b). The second group consisted of accessions mainly from both Konkomba and Anufor ethnic groups (Fig. 2a). The second dimension of the plot (4.9%) separated this second group in two sub-groups: accessions mainly grown by Konkomba farmers, predominantly the Ipui farmer cultivar, and accessions from Anufor farmers, predominantly the Namba farmer cultivar (Fig. 2b).
Principal component analysis carried out on the allelic matrix of the 140 white fonio individuals successfully genotyped with 14 microsatellites markers and projected on the first and second components. a Individuals are differentiated according to the farmer’s ethnicity. b Individuals are colored according to the farmers’ cultivar. All the farmers’ cultivars bearing Ipui names were grouped together and also for Namba. Farmer’s cultivars that were minor were grouped together as Other and colored in grey
Population structure analysis
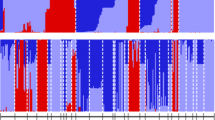
Population structure analysis using sparse non-negative matrix factorization method revealed an optimized sub-structured of K = 6 subpopulations (Fig S2). The Discriminant analysis of principal components (DAPC) also revealed the optimal number of K clusters to be six (lowest Bayesian Information Criterion, BIC after retaining 50 principal components and 4 linear discriminants) (Fig S2 C and D). The Chi2 test with 2000 iterations and the Monte-Carlo test revealed significant (p value = 0.0004) association between the two clustering methods and confirmed the population structure of the 140 fonio accessions. The bar plots from K = 2 to K = 6 revealed that clustering of the accessions was mainly associated with genetic attributes of the accessions rather than the geographical location (Fig. 3).
At K = 2, one subpopulation (pink color) was made up of 50 members with 50% of them collected from farmers belonging to the Anufor ethnic group. Farmers belonging to the Konkomba ethnic group supplied 34% of the accessions while Kabre and Dagomba farmers provided 10% and 6% of the accessions respectively. Majority (72%) of the accessions in this subpopulation were early maturing while 28% of them were late maturing. A mix of farmers’ cultivars composed this group. Subpopulation two (blue color) consisted of 90 accessions of which majority (83.4%) were associated with farmers belonging to the Anufor ethnic group cultivating mostly the Yadema farmer’s cultivar, with early maturing cycle (Fig S5). Other accessions of this group belonged to Konkomba (11.1%) and Kabre (4.4%) ethnic groups and are all early maturing. Only 1.1% of accessions in this subpopulation was collected from a farmer belonging to the Dagomba ethnic group.
At K = 3, the second subpopulation (90 accessions, blue color) at K = 2 sub divided mainly into two groups. The first group consisted of 33 accessions (orange color) of which 79% were associated with early maturity while 21% were late maturing cultivars. With the exception of one accession from the Dagomba ethnic group, all the late maturing accessions in this subpopulation were associated with farmers belonging to the Anufor ethnic group. The second group had the highest number of individuals (60 accessions, blue color). Similarly, 75% of these accessions were early maturing while 25% were late maturing.
The pink cluster at K = 3 with 47 accessions largely divided into two clusters at K = 4. The first subpopulation (pink color) was composed of 20 individuals most of which were early maturing and associated with Anufor farmers cultivating the Namba cultivar. The second subpopulation (green color) had a total of 24 accessions. However, this subpopulation was composed of individuals with mixed maturity classes and majority of these accessions were associated with Konkomba farmers cultivating the Ipui cultivar.
At K = 5, the most significant is the division of the cluster with blue bar plots (67 accessions) at K = 4 into two to form the two largest subpopulations, blue and orange colored bar plots. It is also worthy to note the appearance of the cluster with the sky-blue bar plots with majority of them associated with Anufor farmers. All but one of these accessions were early maturing.
The fonio accessions subdivided into six ancestral populations at K = 6. To account for admixtures and determine the actual number of individuals within the six ancestral populations, a membership coefficient of ≥ 0.70 was applied at K = 6. Sixty-four accessions constituting 45% of the total population were classified as admixed (Table S5). Subpopulation 1 consisted of three early maturing accessions (yellow bar plots). Subpopulation 2 had the largest number of accessions (28, blue bar plots) most of which were associated with Anufor farmers cultivating the Yadema farmers’ cultivar. Subpopulation 3 comprised 25 accessions (orange bar plots) which predominantly correspond to the Yadema farmers’ cultivar. Subpopulation 4 consisted of seven accessions (pink bar plots). These accessions were associated with farmers belonging to Anufor and Kabre ethnic groups cultivating the Namba farmer cultivar. Subpopulation 5 was made up of two accessions (sky blue bar plots) collected from farmers belonging to the Anufor ethnic group. In the case of subpopulation 6, majority of the accessions (green bar plots) were collected from Konkomba farmers cultivating the Ipui cultivar with heterogeneous maturity classes.
There was no significant association between the six clusters inferred at K = 6 and maturity (Chi2 test, p value = 0.40). However, there was significant association with the ethnicity of the farmers (Chi2 test, p value < 0.05) and the farmers' cultivar (Chi2 test, p value < 0.001). When conducting the hierarchical analysis of molecular variance (AMOVA) with ethnicity and farmers' cultivar within ethnicity, 2.80% of the genetic variation was due to the difference between ethnicity, while 16.83% was due to the farmers’ cultivar within ethnicity (16.40% without taking account of ethnicity). A large variation pertained between samples within farmers’ cultivar with 58.52%.
Hierarchical cluster analysis for Anufor ethnicity
Focusing on the main ethnic group cultivating fonio in Ghana, the farmers’ cultivar accounted for 17.52% of the genetic variation, while 56.67% of the variation remained between samples within farmers’ cultivar (AMOVA). The neighbor-joining tree (unrooted) identified three main groups predominantly differentiating the two main cultivars cultivated by the Anufor: Namba and Yadema (Fig. 4).
Isolation by distance (IBD)
There was no IBD pattern detected between communities (Fig. 5a, Mantel tests, p = 0.48, R = − 0.05). The correlation between genetic differentiation (FST)/(1- FST) and geographical distance is presented in Fig. 5b. From the plot, the highest density of comparison among the fonio accessions was within a 20 km radius (Fig. 5b). This shows that most of the accessions were collected in communities within districts less than 20 kms apart.
Isolation by distance analysis (IBD) between the 24 communities producing fonio. a Histogram of permuted values obtained with 1000 Mantel tests assessing correlation between geographical distance and genetic distance under absence of spatial structure. The dot represents the actual value of the correlation. b IBD plot showing the relationship between genetic distance and geographical distance of fonio accessions. The dashed red line represents a linear regression
Genetic diversity of Ghanaian fonio germplasm in relation to the West African fonio diversity
To relate the genetic diversity among the Ghanaian fonio accessions to the global diversity in West Africa, a principal component analysis was performed. The first two principal component (PC) axes accounted for 7.07% of the total variation (Fig. 6). From PC1, fonio accessions from Togo, Nigeria, Mali and Guinea clustered separately from Ghanaian accessions. The Ghanaian accessions separated into two with one group clustering with accessions from other West African countries while the other group mainly accessions from the Chereponi district clustered alone. These accessions with PC1 values greater than 3% were nearly all homozygous (93%) for the De_36 marker with size 186 bp. This allele was almost exclusively present in Ghana, although it was found four times in Benin and Togo. The second PC axis separated accessions from Mali and those from Ghana, Nigeria, Togo and Benin. PC 3 axis accounted for 2.37% of the total variability among the accessions (Fig S6). This axis separated the Ghanaian accessions collected from Chereponi district from those of Benin and Guinea.
Discussion
This research aimed to describe in-situ fonio genetic diversity in Ghana. It paid attention to both biological and social factors that could have influenced over time the organization of fonio genetic diversity. Focusing on the local scale, emphasis was put on the role the Ghanaian farmers have played in maintaining fonio diversity by cultivating and producing their own cultivars season after season.
Social footprint on fonio genetic diversity and structure
In the studied area, 21 farmers’ fonio cultivars were cultivated mainly for household consumption. Each farmers’ cultivar belonged almost exclusively to one ethnic group, and the source of seeds for planting was mainly obtained from the household, i.e. farmer saved seeds. The diversity statistics observed in the research are lower to those of Diop et al. (2023) who reported a total of 132 alleles with a mean of 9.43 alleles per locus, mean observed heterozygosity of 0.05 and expected heterozygosity of 0.43 among 158 Senegalese fonio accessions using SSR markers. The higher average number of alleles per locus and mean expected heterozygosity from their research compared to our findings could be due to the higher number of fonio accessions used and the wider geographical distribution area of fonio in Senegal. The level of allelic richness among the ethnic groups observed in this research is similar to results obtained by Diop et al. (2023) who detected 1.49 to 2.01 unique alleles among eight ethnic groups in Senegal.
Crop genetic diversity has been found to be influenced by human activities such as selection and dissemination of desirable cultivars and long-term interaction with environmental factors (Leclerc and Coppens d’Eeckenbrugge 2011). These factors interact at different scales to shape the genetic diversity observed in most crop species. Fonio in-situ varietal diversity is socially organized in Ghana, and this directly influenced its genetic diversity organization. Indeed, since each farmers’ cultivar belonged almost exclusively to one ethnic group, fonio cultivar can be considered as an identity mark, or at least this suggests the existence of a social barrier between groups. This suggests that seed exchanges among communities are not frequent. This observation is relevant with the farmer-saved seed practices, and with the fonio genetic variability observed between ethnic groups (8.8%), and between cultivars (16.4%). Because fonio is an autogamous crop, the large proportion of seeds that is self produced by farmers imply that genetic diversity within and between cultivars remain the same season after season. Farmer-saved seed practices in fact induce interethnic variability of a crop as a result of selection and limited seed mediated gene flow between ethnic groups (Leclerc and Coppens d’Eeckenbrugge 2011).
Population structure analysis tended to confirm this hypothesis. The FST calculated between the ethnic groups revealed significant differences between some of the ethnic groups (Table S8). For instance, the FST calculated between the Anufor and Konkomba ethnic groups was significant (FST = 0.079, p < 0.001) while the FST between Anufor and Kabre ethnic groups was not significant (FST = 0.028, p > 0.05). Furthermore, structure analysis among the 140 Ghanaian fonio accessions revealed six ancestral populations. A higher number of the fonio accessions (64 individuals) in this research was classified as admixed at an ancestry coefficient of 70%. The presence of an admixed population could be an indication of gene flow among the ancestral populations. The admixed population comprised fonio accessions from all the four ethnic groups. A greater number of the admixed population were collected from the Anufor farmers. But this could be as a result of the higher number of accessions collected from this ethnic group. Although significant FST was observed among some of the ethnic groups, the social organization of fonio genetic diversity in Ghana was largely influenced by farmers’ cultivars, which have contributed to maintaining fonio genetic diversity over time. This observation is relevant with Diop et al. (2023) who found that the genetic diversity of fonio in Senegal was influenced by the ethnicity of farmers. The FST calculated between the Mande and Atlantic families was low but significant (FST = 0.031, p = 0.001). They observed that Fulani people cultivated early maturing accessions while late maturing accessions were cultivated by farmers belonging to the Balante ethnic group. Other research have also reported the influence of ethnicity on genetic diversity patterns of crops such as sorghum (Labeyrie et al. 2014), maize (Orozco-Ramírez et al. 2016) and pearl millet (Naino Jika et al. 2017).
Other social factors such as age, gender and educational level of farmers were not directly linked to the genetic diversity observed in this research as well as biological factors such as maturity period of the fonio accessions. This was evident in the PCA analysis where both early and late maturing fonio accessions clustered together irrespective of the community of collection and ethnicity of the affiliated farmers. In addition, there was no significant isolation by distance among the Ghanaian fonio accessions. This result is contrary to the observation of Adoukonou-Sagbadja et al. (2007) who reported clustering of white fonio accessions based on geographic distance. As there was no isolation by distance, some fonio accessions clustered together irrespective of the geographical locations from where they were collected. Again, some accessions collected from different communities were identified to be duplicates based on the SSR markers used. This could be associated with the recent commercial fonio production characterized by provision of seeds and other incentives to farmers who produce and sell to the companies. For example, fonio cultivation in the Gushegu district of the Northern Region is a recent phenomenon through a company and the crop is solely cultivated for sale to the company.
Challenges and opportunities for conservation and utilization of fonio genetic diversity
This research revealed significant genetic variability among fonio cultivars cultivated by farmers in Ghana, which is unique even more considering the restricted area of fonio cultivation in Ghana. Notably, farmer-held fonio cultivars played a substantial role in shaping the genetic diversity of fonio in Ghana. The findings underscore the pivotal role of farmers’ practices in preserving fonio genetic diversity over time. Moreover, the astonishing distinctiveness of Ghanaian fonio accessions, particularly those from the Chereponi district, stands out in comparison to the global diversity observed in West African collections. Although the fonio accessions from Ghana are localized and close to Togo, there is a unique diversity in the Ghanaian accessions. To understand the factors contributing to this unique diversity, future research should prioritize an extensive ethnobotanical investigation. This would help unravel the processes behind the selection of farmers’ fonio cultivars and shed light on the social and environmental factors influencing fonio genetic diversity in Ghana. Recognizing the significance of social factors in shaping fonio genetic diversity, strategies for the management and conservation of fonio genetic resources should consciously incorporate traditional farming practices. This holistic approach acknowledges the symbiotic relationship between fonio, farmers, and their practices, ensuring the preservation of fonio genetic diversity while considering the social and environmental contexts in which it thrives.
The observed genetic diversity among the different populations (community, ethnicity and farmers’ cultivar) offers an opportunity for targeted exploitation through the identification and selection of genetically distinct individuals, laying the foundation for tailored product profiles within fonio breeding programs to cater to diverse market segments. The admixture population with a wider genetic base may have broad adaptability and greater resilience to the changing climate which could be harnessed in breeding programmes. To this, there is the need to evaluate the fonio accessions for their performance in traits of agronomic importance to aid in the selection of desirable parental genotypes for crop improvement purposes. Since the diversity of Ghanaian accessions is truly original, we also expect it to contribute to a certain originality of the agronomic traits studied. Additionally, genotyping by sequencing could provide an insight into genomic regions associated with useful agronomic traits to identify superior genotypes. However, the highly autogamous nature of fonio coupled with its small-sized flowers requires that efficient hybridization strategies are identified to develop new fonio cultivars that will meet the needs of farmers and other end users. Targeted breeding programmes could focus on improving traits such as earliness, lodging, shattering, seed size and grain yield.
Conclusion
Unique genetic diversity was detected among the Ghanaian fonio accessions, with farmers’ practices and knowledge playing a significant role in organizing this diversity over time. Future research should prioritize extensive ethnobotanical analyses to unravel the factors contributing to the pattern of fonio genetic diversity. Integrating traditional farming practices into conservation and breeding strategies is essential for developing tailored fonio cultivars suited to diverse market needs and environmental changes. This is even more important as Ghana’s peculiar diversity suggests specific features of interest to farmers.
Data availability
The data from the farmer surveys and genotyping were used in drafting the manuscript.
References
Abrouk M, Ahmed HI, Cubry P, Šimoníková D, Cauet S, Pailles Y, Bettgenhaeuser J et al (2020) Fonio millet genome unlocks African orphan crop diversity for agriculture in a changing climate. Nat Commun 11(1):4488. https://doi.org/10.1038/s41467-020-18329-4
Addai IK, Bisuki BK, Bawa A (2022) Evaluation of fonio for adaptive traits. Adv Agric 2022:1–10. https://doi.org/10.1155/2022/4194746
Adjebeng-Danquah J, Manu-Aduening J, Asante IK, Agyare RY, Gracen V, Offei SK (2020) Genetic diversity and population structure analysis of Ghanaian and exotic cassava accessions using simple sequence repeat (SSR) markers. Heliyon 6(1):e03154. https://doi.org/10.1016/j.heliyon.2019.e03154
Adoukonou-Sagbadja H, Dansi A, Vodouhè R, Akpagana K (2006) Indigenous knowledge and traditional conservation of fonio millet (Digitaria exilis, Digitaria iburua) in Togo. Biodivers Conserv 15(8):2379–2395. https://doi.org/10.1007/s10531-004-2938-3
Adoukonou-Sagbadja H, Wagner C, Dansi A, Ahlemeyer J, Daïnou O, Akpagana K, Ordon F, Friedt W (2007) Genetic diversity and population differentiation of traditional fonio millet (Digitaria Spp.) landraces from different agro-ecological zones of West Africa. Theor Appl Genet 115(7):917–931. https://doi.org/10.1007/s00122-007-0618-x
Barnaud A, Vignes H, Risterucci A-M, Noyer J-L, Pham J-L, Blay C, Buiron M, Vigouroux Y, Billot C (2012) Development of nuclear microsatellite markers for the fonio, Digitaria exilis (Poaceae), an understudied West African cereal. Am J Bot 99(3):e105–e107. https://doi.org/10.3732/ajb.1100423
Bengtsson H, Ahlmann-Eltze C, Bravo H.C, Gentleman R, Gleixner J (2023) matrixStats: functions that apply to rows and columns of matrices (and to vectors)
Botstein D, White RL, Skalnick MH, Davies RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphism. Am J Hum Genet 32:314–331
Clottey VA, Agyare WA, Bayorbor TB, Abanga JJ, Kombiok JM (2006a) Genetic relatedness of fonio (Digitaria Spp.) landraces assembled in Ghana. Plant Genet Resour Newsl 147:6–11
Clottey VA, Agyare WA, Kombiok JM, Abdulai H, Kaleem AH (2006b) Fonio (Digitaria Spp.) germplasm assemblage for characterization, conservation and improvement in Ghana. Plant Genet Resour Newsl 146:24–27
Delêtre M, McKey DB, Hodkinson TR (2011) Marriage exchanges, seed exchanges, and the dynamics of Manioc diversity. Proc Natl Acad Sci 108(45):18249–18254. https://doi.org/10.1073/pnas.110625910
Diop BM, Gueye MC, Agbangba CE, Cisse N, Deu M, Diack O, Fofana A et al (2018) Fonio (Digitaria exilis (Kippist) Stapf): a socially embedded cereal for food and nutrition security in Senegal. Ethnobiol Lett 9(2):150–165. https://doi.org/10.14237/ebl.9.2.2018.1072
Diop BM, Guèye MC, Leclerc C, Deu M, Zekraoui L, Calatayud C, Rivallan R et al (2023) Ethnolinguistic and genetic diversity of fonio (Digitaria exilis) in Senegal. Plants People Planet. https://doi.org/10.1002/ppp3.10428
Dray S, Dufour A-B (2007) The Ade4 package: implementing the duality diagram for ecologists. J Stat Softw 22(4):1–20. https://doi.org/10.18637/jss.v022.i04
Frichot E, Francois O (2015) LEA: an R package for landscape and ecological association studies. Methods in ecology and evolution. Bioconductor. https://doi.org/10.18129/B9.BIOC.LEA
Goudet J, Jombart T (2022) hierfstat: Estimation and Tests of Hierarchical F-Statistics. R package version 0.5-11. https://CRAN.R-project.org/package=hierfstat
Govindaraj M, Vetriventhan M, Srinivasan M (2015) Importance of genetic diversity assessment in crop plants and its recent advances: an overview of its analytical perspectives. Genet Res Int. https://doi.org/10.1155/2015/431487
Jideani IA, Jideani VA (2011) Developments on the cereal grains Digitaria exilis (Acha) and Digitaria iburua (Iburu). J Food Sci Technol 48(3):251–259. https://doi.org/10.1007/s13197-010-0208-9
Jombart T (2008) Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24(11):1403–1405. https://doi.org/10.1093/bioinformatics/btn129
Jombart T, Devillard S, Balloux F (2010) Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet 11(1):94. https://doi.org/10.1186/1471-2156-11-94
Kaczmarek T, Causse S, Abdul SD, Abraham S, Achigan-Dako EG, Adje C, Adjebeng-Danquah J et al (2023) Towards conservation and sustainable use of an indigenous crop: a large partnership network enabled the genetic diversity assessment of 1539 fonio (Digitaria exilis) accessions. Plants People Planet. https://doi.org/10.1002/ppp3.10424
Kamvar ZN, Tabima JF, Grünwald NJ (2014) Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2:e281. https://doi.org/10.7717/peerj.281
Kassambara A, Mundt F (2020) Factoextra: extract and visualize the results of multivariate data analysis. https: CRAN. R-project.org/package=factoextra
Labeyrie V, Deu M, Barnaud A, Calatayud C, Buiron M, Wambugu P, Manel S, Glaszmann J-C, Leclerc C (2014) Influence of ethnolinguistic diversity on the sorghum genetic patterns in subsistence farming systems in Eastern Kenya. Edited by John P. Hart. PLoS ONE 9(3):e92178. https://doi.org/10.1371/journal.pone.0092178
Leclerc C, Coppens d’Eeckenbrugge G (2011) Social organization of crop genetic diversity. The G × E × S interaction model. Diversity 4(1):1–32. https://doi.org/10.3390/d4010001
Naino Jika AK, Dussert Y, Raimond C, Garine E, Luxereau A, Takvorian N, Djermakoye RS, Adam T, Robert T (2017) Unexpected pattern of pearl millet genetic diversity among ethno-linguistic groups in the Lake Chad Basin. Heredity 118(5):491–502. https://doi.org/10.1038/hdy.2016.128
Orozco-Ramírez Q, Ross-Ibarra J, Santacruz-Varela A, Brush S (2016) Maize diversity associated with social origin and environmental variation in Southern Mexico. Heredity 116(5):477–484. https://doi.org/10.1038/hdy.2016.10
Padgham M (2021) Geodist: fast, dependency-free geodesic distance calculations. R Package version 0.0.7. https://github.com/hypertidy/geodist
Paradis E (2010) Pegas: an R package for population genetics with an integrated–modular approach. Bioinformatics 26(3):419–420. https://doi.org/10.1093/bioinformatics/btp696
R Core Team (2022) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria
Risterucci A-M, Hippolyte I, Perrier X, Xia L, Caig V, Evers M, Huttner E, Kilian A, Glaszmann J-C (2009) Development and assessment of diversity arrays technology for high-throughput DNA analyses in Musa. Theor Appl Genet 119(6):1093–1103. https://doi.org/10.1007/s00122-009-1111-5
Rousset F (1997) Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 145(4):1219–1228. https://doi.org/10.1093/genetics/145.4.1219
Stevens A, Ramirez-Lopez L (2022) An introduction to the prospectr package. R package Vignette. R version 0.2.6
Venables WN, Ripley BD (2010) Modern applied statistics with S. Statistics and computing, 4th edn. Springer, New York
Vodouhe R, Dako GA, Dansi A, Adoukonou-Sagbadja H (2007) Plant genetic resources and food security in west and central Africa. Bioversity International, Rome
Wickham H (2016) Ggplot2: elegant graphics for data analysis. Springer, New York. https://doi.org/10.1007/978-3-319-24277-4
Yu G (2022) Data integration, manipulation and visualization of phylogenetic trees, 1st edn. Chapman and Hall, Boca Raton. https://doi.org/10.1201/9781003279242
Acknowledgements
This research was funded by the EU H2020 EWA-BELT project [862848] “Linking East and West African farming systems experience into a BELT of sustainable intensification” through University of Sassari, Italy. We are grateful to the community leaders, farmers and staff of the Department of Agriculture, in the Chereponi, Gushegu, Saboba, Tatale and Zabzugu districts, for their support and active participation.
Funding
This research was funded by the EU H2020 EWA-BELT project [862848] “Linking East and West African farming systems experience into a BELT of sustainable intensification”.
Author information
Authors and Affiliations
Contributions
RYA, JA-D, CB, AB, AWK and RA designed the experiment; AB and CB carried out the training on germplasm expedition and collection; RYA, JA-D, DKP, TKT, JMK and SKB collected the fonio accessions and associated ethnobotanical data; SC genotyped the samples; RYA, JA-D, TK, TG, CL, AB and CB analyzed the data; RYA, TK, CL, JA-D, AB, CB, AWK and RA wrote the manuscript; all authors revised the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest exists.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Agyare, R.Y., Kaczmarek, T., Leclerc, C. et al. Revealing Ghana’s unique fonio genetic diversity: leveraging farmers knowledge for sustainable conservation and breeding strategies. Genet Resour Crop Evol (2024). https://doi.org/10.1007/s10722-024-01974-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10722-024-01974-9