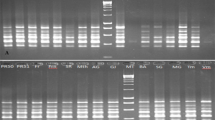
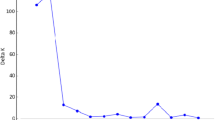
Abstract
Prunus salicina 'Shazikongxinli' is one of China’s most economically valuable and reputable Prunus salicina cultivars. Understanding the genetic diversity and population structure of 'Shazikongxinli' is imperative for excellent germplasm breeding and the preservation of genetic resources. This work used morphological markers, inter-retrotransposon amplified polymorphism (IRAP), and inter simple sequence repeat (ISSR) to assess the genetic variation status of 50 'Shazikongxinli', with 18 plum cultivars selected as the control outgroup. The results showed that the average Shannon–Weaver diversity index (H') of 32 qualitative traits in 50 tests 'Shazikongxinli' was 0.557, the average coefficient of variation of 11 quantitative traits was 15.57%, and a total of 232 and 105 polymorphic loci were obtained from 22 IRAP and 15 ISSR primers, respectively. All three marker systems showed relatively rich polymorphism, especially the IRAP markers. This might be related to the nature of the retrotransposons in the IRAP markers, which might be more suitable for intraspecific variability detection than ISSR. In addition, all three markers clustered the 68 tested germplasms into two groups. All of 'Shazikongxinli' clustered into one group, and most outgroup plum cultivars clustered into another group. This suggested a relatively narrow genetic base within the 'Shazikongxinli' population. These results would be useful in understanding the genetic diversity of the germplasm resources of 'Shazikongxinli' and provide a basic theoretical reference for breeding superior germplasm.




Similar content being viewed by others
References
Acuna CV, Rivas JG, Brambilla SM et al (2019) Characterization of genetic diversity in accessions of Prunus salicina Lindl: keeping fruit flesh color ideotype while adapting to water stressed environments. Agronomy 19(9):487
Almajali D, Abdel AH, Migdadi H (2012) Evaluation of genetic diversity among Jordanian fig germplasm accessions by morphological traits and ISSR markers. Sci Hortic 147:8–19
Baslilio C, Carole D, Mario M et al (2012) Genetic characterization of Japanese plum cultivars (Prunus salicina) using SSR and ISSR molecular markers. Cienc Investig Agrar 39(3):533–543
Ben TH, Ben MS, Baraket G et al (2015) Assessment of genetic diversity and relationships among wild and cultivated Tunisian plums (Prunus spp.) using random amplified microsatellite polymorphism markers. Genet Mol Res 14:1942–1956
Brenda I, Guerrero MEG et al (2022) Simple sequence repeat (SSR)-based genetic diversity in interspecific plumcot-type (Prunus salicina × Prunus armeniaca) hybrids. Plants (basel) 11(9):1241
Goulao L, Monte C, Oliveira CM (2001) Phenetic characterization of plum cultivars by high multiplex ratio markers: amplified fragment length polymorphisms and inter-simple sequence repeats. J Am Soc Hortic Sci 126:72–77
Guan CF, Chachat S, Zhang PX et al (2020) Inter- and intra-specific genetic diversity in diospyros using SCoT and IRAP markers. Hortic Plant J 6(2):71–80
Hu GM, Li CB, Yang B et al (2022) Analysis and comprehensive evaluation of fruit trait diversity of 72 Actinidia chinensis in Yichang. J Fruit Sci 39(9):1540–1552
Kalendar R, Alan HS (2014) Transposon-based tagging: IRAP, REMAP and iPBS. Methods Mol Biol 1115:233–255
Kalendar R, Grob T, Regina M et al (1999) IRAP and REMAP: two new retrotransposon-based DNA fingerprinting techniques. Theor Appl Genet 98:704–711
Karim S, Mehrana KD, Sezai E et al (2017) Comparison of traditional and new generation DNA markers declares high genetic diversity and differentiated population structure of wild almond species. Sci Rep 7(1):5966
Kazija DH, Jelacic T, Vujevic P et al (2014) Plum germplasm in croatia and neighboring countries assessed by microsatellites and DUS descriptors. Tree Genet Genom 10:761–778
Li L, Wang HY, Tao SY et al (2011) Cluster analysis of plum germplasm resources based on fruit quality traits and SSR markers. J Hunan Agric Univ 46(1):38–47
Liu W, Liu D, Zhang A et al (2007) Genetic diversity and phylogenetic relationships among plum germplasm resources in China assessed with inter-simple sequence repeat markers. J Am Soc Hortic Sci 132(5):619–628
Liu D, He X, Liu G et al (2011) Genetic diversity and phylogenetic relationship of Tadehagi in southwest China evaluated by inter-simple sequence repeat (ISSR). Genet Resour Crop Evol 58:679–688
Lynch M, Pfrender M, Spitze K et al (1999) The quantitative and molecular genetic architecture of a sub-divided species. Evolution 53:100–110
Martínez GP, Arulsekar S, Potter D et al (2003) Relationships among peach and almond and related species as detected by SSR markers. J Am Soc Hortic Sci 128:667–671
Mei LN, Wen XP, Fan FH et al (2021) Genetic diversity and population structure of masson pine. (Pinus massoniana Lamb.) superior clones in South China as revealed by EST-SSR markers. Genet Resour Crop Evol 68:1987–2002
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci USA 70:3321–3324
Pop R, Harţa M, Szabo K et al (2018) Genetic diversity and population structure of plum accessions from a Romanian germplasm collection assessed by simple sequence repeat (SSR) markers. Notulae Botanicae Horti Agrobotanici Cluj-Napoca 46(1):90–96
Rana JC, Chahota RK, Sharma V et al (2015) Genetic diversity and structure of Pyrus accessions of Indian Himalayan region based on morphological and SSR markers. Tree Genet Genom 11(1):821
Rohlf FJ (2000) NTSYS-PC, Numerical taxonomy and multivariate analysis system, version 2.1. Exeter Publications, New York
Ruchi T, Vinay S, Amish KS et al (2020) Genetic diversity and population structure detection in sponge gourd (Luffa cylindrica) using ISSR, SCoT and morphological markers. Physiol Mol Biol Plants 26(1):119–131
Sehic J, Nybom H, Hjeltnes SH et al (2015) Genetic diversity and structure of Nordic plum germplasm preserved ex situ and on-farm. Sci Hortic 190:195–202
Shannon C, Weaver W (1949) The mathematical theory of communication. University of Illinois Press, Urbana
Tao J, Qiao G, Wen XP et al (2014) Characterization of genetic relationship of dragon fruit accessions (Hylocereus spp.) by morphological traits and ISSR markers. Sci Hortic 170:82–88
Urrestarazu J, Errea P, Miranda C et al (2018) Genetic diversity of Spanish Prunus domestica L. germplasm reveals a complex genetic structure underlying. PLoS ONE 13:1–21
Wang XM, Wu WL, Zhang CH et al (2015) Analysis of the genetic diversity of beach plums by simple sequence repeat markers. Genet Mol Res 14(3):9693–9702
Wei X, Shen F, Zhang QP et al (2021) Genetic diversity analysis of Chinese plum (Prunus salicina L.) based on whole-genome resequencing. Tree Genet Genom 17:26
Wu W, Chen F, Yeh K et al (2019) ISSR analysis of genetic diversity and structure of plum varieties cultivated in Southern China. Biology (basel) 8:1–13
Wu MH, Wang G, Wang LL et al (2022) Genetic diversity evaluation of Prunus salicina Lindl. ‘Shazi kongxinli’ based on morphological and ISSR marker. Mol Plant Breed 9:487
Yao ZF, Zhang XJ, Yang YL et al (2022) Genetic diversity of phenotypic traits in 177 sweetpotato landrace. Acta Agron Sin 48(9):2228–2241
Yeh FC, Boyle TJB (1997) Population genetic analysis of codominant and dominant markers and quantitative traits. Belg J Bot 129:157
Yu X, Zhang Q, Liu W et al (2011) Genetic diversity analysis of morphological and agronomic characters of Chinese plum (Prunus salicina Lindl.) germplasm. J Plant Genet Resour 12(3):402–440
Zhang J, Zhou E (1998) China fruit-plant monographs, plum flora. China Forestry Press, Beijing
Zhang SS, Wu S, Xiao HY et al (2020) Nutritional characteristics of Shazikongxinli fruits produced in Yanhe county. Food Ind 41(3):340–343
Funding
The project was supported by grants from the Science and Technology Program of Guizhou Province, China (Grant Numbers [2019] 4013 and [2020] 1Y023).
Author information
Authors and Affiliations
Contributions
QG conceived and designed the experiments. WG and LRR performed the experiments. WG and WLL analyzed the data and wrote the paper. RFH collected samples and managed the materials. WMH obtained the funding. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest. All the authors read and approved the manuscript in its final form.
Data availability
The datasets generated and analyzed during the current study are available from the corresponding author upon reasonable request.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, G., Li, R., Wu, M. et al. Assessment of genetic diversity of Prunus salicina 'Shazikongxinli' by morphological traits and molecular markers. Genet Resour Crop Evol 70, 2727–2739 (2023). https://doi.org/10.1007/s10722-023-01599-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-023-01599-4