Abstract
Simple sequence repeat (SSR) microsatellite markers have been extensively used to identify duplication and analyse genetic diversity in germplasm collections of apple. Here, we present findings from the use of a standard set of SSR loci in the managed repropagation of a significant international germplasm collection: the UK National Fruit Collection (NFC). A subset of eight SSR loci was deemed sufficient to distinguish all apart from the clonal relatives across a sample of 1995 accessions, with a single exception being one pair of full siblings. In total, 99% of accessions were able to be confirmed present and correct after the replacement of trees initially identified to be incorrectly propagated. In parallel to the curation of the collection itself, through an initiative led by the UK local apple enthusiast community, 3373 SSR profiles for apples held in local collections were compared to the NFC holdings. Overall, in both sets of material, diversity remained high with average gene diversity values of 0.800 and 0.812 in the NFC holdings and local collections, respectively. Accessions in local collections were not found to differ in their overall coverage of genetic diversity to that of the NFC collection (FST = 0.0035) although significant numbers of locally valued, and genetically distinguishable individuals were identified, some of which may represent ‘lost’ cultivars.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Domesticated apple (Malus x domestica Borkh.) is one of the most important temperate fruit crops in the world. Cultivars of apple are propagated vegetatively, typically by grafting, and field collections of clonal material and seedlings are held in many countries where production is based (Bramel and Volk 2019). Within the UK, cultivated apple is maintained, at a national level, within the National Fruit Collection (NFC) owned by the Department for Environment, Food and Rural Affairs and curated by the University of Reading at Brogdale Farm in Kent. As in many countries with a history of apple growing, the UK has a number of non-governmental organisations (NGOs) that have an interest in the local history of apple cultivars, and a number of these NGOs maintain collections of locally important material. Private individuals also maintain collections of apples for a range of purposes, from small-scale production to research and conservation. Many of these local collectors use the NFC as their reference point for cultivar identification.
Microsatellite markers, also known as simple sequence repeat markers (SSRs), have been available in apple for many years (Guilford et al. 1997) and their utility for the assessment of germplasm has been tested on numerous occasions. Early studies (Hokanson et al. 1998) established that the technology was able to differentiate material in collections. Numerous subsequent studies have utilised SSR markers to screen collections for duplication and diversity (Ferreira et al. 2016; Garkava-Gustavsson et al. 2008, 2013; Gasi et al. 2016; Gasi et al. 2010; Gharghani et al. 2009; Gross et al. 2014, 2012; Guarino et al. 2006; Marconi et al. 2018; Patzak et al. 2012; Pereira-Lorenzo et al. 2007; Pereira-Lorenzo et al. 2017; Testolin et al. 2019; van Treuren et al. 2010) and to identify core collections and subsets (Kim et al. 2019; Lassois et al. 2016; Liang et al. 2015; Pereira-Lorenzo et al. 2018), mostly on a national basis. Further studies have used the technology to examine parentage in pedigreed material (Cabe et al. 2005; Evans et al. 2011; Kitahara et al. 2005; Lassois et al. 2016; Moriya et al. 2011; Salvi et al. 2014) and compare the genetic diversity held across larger regions such as Europe (Urrestarazu et al. 2016). An original SSR dataset for the UK NFC was initially developed by Fernandez–Fernandez (2010) and updated during subsequent curation of the collection (Ordidge and Venison 2018). The NFC dataset was based on the first priority group of loci proposed by the European Cooperative Programme for Plant Genetic Resources (ECPGR) Fruit Network (Fernandez-Fernandez 2013). This recommended set of loci was developed with the view of allowing collaboration between collections through the alignment of data. Recent efforts have attempted to align SSR data from multiple collections within a network of European researchers; a Malus UNiQue genotype code (MUNQ) as a development from the FBUNQ code described by Urrestarazu et al. (2016) has been allocated to a number of genotypic profiles of apple (Denancé et al. 2020; Muranty et al. 2020).
For reasons of efficiency and ease of management, many field collections of apple, similar to commercial orchards, are maintained on dwarfing rootstocks. It is generally accepted that dwarfing stocks reduce the longevity of trees, although an age of approximately 50 years on ‘M9’ rootstock is not uncommon. Consequently there is a need for field collections to be cyclically repropagated.
The apple collection at the NFC has been through a number of repropagation events in recent times, beginning when the collection was relocated and established at Brogdale Farm in the early 1950s. This collection (NFC1) was then repropagated and planted out as NFC2 during the years 1975–1978 before the most recent repropagation event, which has recently been completed as NFC3.
Inevitably, through the collection of dormant graftwood, the grafting and maintenance of trees in a nursery and the planting out of the newly repropagated trees, there are opportunities for error and mislabelling. Genetic markers offer an opportunity to improve grafting verification, from the basis of comparing material, often fruit, at a morphological level; they are not affected by the age of tree or micro-environmental factors and can be applied on trees that are younger than those reliably producing fruit for comparison. Both microsatellite markers (Irish et al. 2014) and single nucleotide polymorphisms (Olasupo et al. 2018) have been used previously to identify propagation errors during the management of collections of similarly vegetatively propagated crops.
Collections of apples, and other fruit, held by NGOs have been aligned as part of national conservation strategies in some countries (Flachowsky 2011; Kellerhals and Egger 2004). The potential for national collections of germplasm to be complemented by collections of material held by NGOs (Lassois et al. 2016; Marconi et al. 2018; van Treuren et al. 2010), farmers (Pereira-Lorenzo et al. 2008) and private individuals (Gross et al. 2018; Magby et al. 2019; Routson et al. 2009) as well as the potential to identify significant trees in historic orchards on public land (Volk et al. 2021) has been investigated with the aid of SSR analysis through a number of research projects. It remains however, less common for these comparisons to be led by the local community itself.
Here, we present the findings of the evaluation of the repropagated NFC3 apple collection using SSR markers. We discuss the ability of this approach to both curate the collection itself and the associated dataset detailing SSR profiles for the individual accessions. We also discuss the findings of a recent initiative, led by the UK apple volunteer community to use microsatellite technology as a method of identifying unknown material and questioning uncertain material in local collections, using the NFC as a reference point (the fruitID [DNA] scheme (fruitID 2021)). We highlight an approach to recognize the genetically distinguishable material held in local (NGO and private) collections through a publicly available Register of Local Cultivars.
Materials and methods
Plant material and DNA extraction
Young leaf material was collected from 3985 trees in the NFC3 apple collection. The samples represented 1997 accessions, the vast majority (1981) of which were each represented by two trees in the collection (this total includes the resampling of seven trees following their replacement after initial analysis). All leaf samples were stored at − 20 °C prior to extraction, which was carried out using the QIAGEN DNeasy 96 Plant Kit according to the manufacturer’s protocol. A series of additional samples were collected in order to either correct sampling errors, check allele size calling queries, confirm repropagation errors or to confirm where new profiles indicated errors in the prior dataset.
Approximately 3400 samples from the local groups were collected as part of the fruitID scheme by individual participants; young leaf material was collected wherever possible. Prior to collection, a sample submission form was completed by each participant and the participant was supplied with a number of pre-labelled collecting envelopes to ensure that all samples in the scheme were sequentially labelled for downstream analysis. DNA extractions were performed at NIAB-EMR using the protocol described by Edge-Garza et al. (2014) with the modification that 5 M sodium chloride was used instead of 6 M ammonium acetate. DNA pellets were re-suspended in 10 mM Tris–HCl pH 8.0 and diluted to 5 ng/µl for use.
PCR amplification and fragment length analysis for NFC samples
For the analysis of NFC3 samples, only the small (CH04c07 [6-FAM], CH01h01 [NED], Hi02c07 [PET], CH01h10 [VIC]) and large (CH04e05 [6-FAM], CH02c11 [NED], CH02c09 [PET], CH02d08 [VIC]) multiplex sets (Fernandez-Fernandez 2013) were initially used. Forward primers were labelled at the 5′ end and small and large multiplex sets were pooled together. Each reaction was carried out in a final volume of 11 µL containing approximately 10 ng genomic DNA and each primer at a concentration of 0.2 µM. The QIAGEN Type-it™ Microsatellite PCR kit (QIAGEN, MD, USA) was used for all reactions according to the manufacturer’s basic instructions (without the addition of Q-solution). In cases where the findings suggested an error in the original dataset (Fernandez-Fernandez 2010), or where prior data were not available, the medium multiplex set (CH01f02 [6-FAM], GD12 [NED], GD147 [PET], CH01f03b [VIC]) (Fernandez-Fernandez 2013) was additionally amplified in order to produce a complete new profile for the accession. All reactions were carried out in an Applied Biosystems Veriti 96-well thermocycler (Applied Biosystems, MA, USA). Amplification was performed as follows: 5 min at 94 °C; 10 cycles of 30 s at 94 °C, 90 s at 60–50 °C reducing by 1 °C per cycle, and 60 s at 72 °C; 25 cycles of 30 s at 94 °C, 90 s at 50 °C and 60 s at 72 °C, with a final stage of 30 min at 60 °C. Following amplification, PCR products were separated on an Applied Biosystems 3730xl capillary sequencer by Source Bioscience (Nottingham, UK) using a LIZ 500 standard.
Fragment analysis was carried out using Genemapper™ 5.0 software (Applied Biosystems, MA, USA). Bins were set on the basis of an initial comparison of a series of samples which were visually aligned with the data in the *.fsa files from the original dataset (Fernandez-Fernandez 2010) on the basis of accession name and tree position. Following this initial comparison, an adjustment factor was placed on each bin, in order to output data such that identical alleles would align with the rounded sizing of the original scores. Subsequent samples were analysed using this bin set and allele calls were compared directly with the original dataset (Fernandez-Fernandez 2010).
PCR amplification and fragment length analysis for fruitID scheme samples
PCR reactions for the fruitID scheme samples were carried out using two multiplexes. Multiplex SL included the previous small (CH04c07 [6-FAM], CH01h01 [NED], Hi02c07 [PET], CH01h10 [HEX]) and large (CH04e05 [6-FAM], CH02c11 [NED], CH02c09 [PET], CH02d08 [HEX]) multiplex sets. Multiplex M included the medium locus set (CH01f02 [6-FAM], GD12 [NED], GD147 [PET], CH01f03b [HEX]) (Fernandez-Fernandez 2013). Amplification was performed in 13 µl volumes using the Type-it™ Microsatellite PCR kit (QIAGEN, MD, USA) according to manufacturer’s protocol, with 0.2 µM of each primer. Thermal cycling was carried out in a Veriti 96 well thermal cycler (Applied Biosystems, MA, USA) as follows: initial 5 min denaturation at 95 °C; 10 touchdown cycles comprising a 30 s denaturation step at 95 °C, followed by 90 s of annealing starting at 55 °C in the first cycle and decreasing 0.5 °C per cycle, and 30 s of extension at 72 °C; 20 cycles of 30 s at 95 °C, 90 s at 50 °C and 30 s at 72 °C; and a final extension step of 30 min at 60 °C. The ECPGR reference genotypes ‘Delicious’, ‘Fiesta’, ‘Malling 9’, ‘Michelin’, ‘Prima’, ‘Worcester Pearmain’, ‘Malus robusta 5’ and ‘Malus floribunda 821’ were included, in earlier analysis such that at least three standards were included in each plate and in later analysis such that at least one standard was present per 16-capillary injection.
Following PCR amplification, products were diluted (1:10) and 1.3 µl separated using an ABI 3130xl Genetic Analyser (Applied Biosystems, MA, USA). Data were collected and alleles sized using Genescan® and Genotyper® software applications (Applied Biosystems, MA, USA). Estimated allele sizes were rounded and harmonized using Excel (MS Office) following automated scoring and sizing in Genotyper® software (Applied Biosystems, MA, USA). Allele sizing was harmonised across plates according to the profiles obtained for the ECPGR standards.
Harmonised allele profiles were compared using GenAlEx software (Peakall and Smouse 2006; 2012) in order to identify matches and near matches (the number of loci used to identify near matches being set at 16 out of 24). Potentially matching profiles were subsequently assessed and verified manually in order to allow for miscalls due to stuttering and null alleles. Profiles were compared to a database of genotypes including both the NFC dataset and unpublished data from previous analysis of collections held by the Irish Seedsavers Association, University College Dublin (both in the Republic of Ireland) and National Trust Cotehele, UK. MUNQ codes were applied following the comparison of genotypes with the existing MUNQ code set (Denancé et al., in prep).
Amalgamation of NFC3 data with prior datasets
Where reproduced allele scores from the small and large multiplex sets were in agreement with the prior dataset (Fernandez-Fernandez 2010; Ordidge and Venison 2018) (allowing for the re-calling of a small number of alleles), the original scores in the prior data for the alleles in the medium multiplex were assumed to be correct and were not reproduced. In cases where the profile was judged to be in disagreement, the medium multiplex was reproduced in order to either confirm or reject the error and, where necessary, produce a complete new profile for the accession. The alleles of the medium multiplex were imputed from what was deemed to be a correctly scored clonal relative in a subset of cases where the original profile in the prior data had been found to be in error and the suggestion of clonality from the reproduced data for the small and large multiplex were supported by previous independent findings (Ordidge et al. 2018). Consequently, a dataset was produced that included an amalgamation of original and new allele scores for the medium multiplex. These data were further amalgamated with prior data (Fernandez-Fernandez 2010; Ordidge and Venison 2018) for a series of accessions that were still in the repropagation process, a small number of accessions that had died since the prior analysis and any accessions that were either not scheduled for, or had been excluded from, repropagation i.e. the NFC cider collection and a series of duplicate accessions that had been marked for deaccession (Online Resource 1; Venison and Ordidge 2022). Again, MUNQ codes were applied following the comparison of genotypes with the existing MUNQ code set (Denancé et al., in prep).
Generation of genetic diversity metrics
NFC data were initially screened to retain 1835 entries distinguishable by allocated MUNQ code (Online Resource 1). fruitID scheme data were screened to retain only entries (982) that were distinguishable from the NFC dataset across any of the 12 loci (Online Resource 2). Diploid, triploid and tetraploid entries (identified by either allele number or prior knowledge) were retained and allele frequencies, gene diversity (Nei 1978) and FST (Weir and Cockerham 1984) were generated using SPAGeDI 1.4 (Hardy and Vekemans 2002). As per Testolin et al. (2019) and following the suggestion of Jamshidi and Jamshidi (2011) data were subsequently converted to a binary format in order to allow the inclusion of polyploid entries within AMOVA (Excoffier et al. 1992) and PCoA which was carried out using GenAlEx 6.503 (Peakall and Smouse 2006, 2012). Following the removal of all accessions with any loci reporting more than two alleles (291 accessions indicating potential triploidy/tetraploidy in the NFC set) a further reduced diploid-only dataset of 1544 accessions from the NFC set only was used to estimate a non-exclusion probability using Cervus 3.07 (Kalinowski, 2007).
Results
SSR data summary for the NFC
In total 3982 SSR profiles representing 1995 accessions were produced from the NFC3 samples (three additional samples, which should have represented two further accessions, were deemed to be sampling errors and data were not analysed further).
Identification of NFC propagation errors
The eight loci in the small and large multiplexes were sufficient to allow the identification of 1849 genotypes (Venison and Ordidge 2022). An additional 16 profiles could be distinguished when considering the full set of 12 loci; however, 14 of these were distinguished only by either null alleles or allele size differences of 1–2 bp (seven profiles each). The majority of these differences was found in loci GD147 and CH01f02 (six and five instances respectively). The remaining two instances included one pair of accessions (2011–042 Reinette du Mans and 1947–143 De Jaune) held under cultivar names believed to be synonymous, suggesting the distinguishing factor to be a potential error in the data, and another where the additional loci distinguished between two accessions (1948–022 Downton Pippin and 2000–102 Yellow Ingestrie) of cultivars documented to be full siblings (Hogg 1884).
Within the group of 1849 genotypes, a further set of 16 instances was identified in which accessions with differing genotypes had been allocated to the same MUNQ code and in all cases the distinguishing elements were either null alleles or allele size differences of 1–2 bp (Online Resource 1; Venison and Ordidge 2022).
Out of the 3982 repropagated trees that were able to be tested, 78 (2%) indicated an initial error in repropagation. In 33 cases the incorrect tree was confirmed to be a ‘Golden Delicious’ interstock, as a result of a failed graft; in 45 cases trees were found to have been mixed up during either repropagation or planting. Following the replacement of the seven trees that were able to be repropagated during the analysis, a total of 1987 of the 1995 accessions (99%) were accepted to have at least one tree present and correct (Online Resource 1). The remaining accessions were either found to be missing as the result of propagation errors in both trees (eight accessions) or could not be tested due to sampling error (two accessions, as above).
Improvement of the dataset through reproduction
Prior scores for the medium multiplex were retained for 1695 accessions. Either a new or confirmatory medium multiplex score was produced for 251 accessions and 25 profiles contained medium multiplex scores imputed from what was deemed to be a clonal relative (Online Resource 1). Following the amalgamation of the newly produced and compiled profiles with the prior data for accessions that had not been repropagated, and the removal of replicate entries from pairs of replicate trees, the total number of profiles in the combined dataset was 2206 (Online Resource 1; Venison and Ordidge 2022). The samples required to produce new medium multiplex scores for 16 accessions were either missed, or failed during the analysis, and with the exception of the three examples described below, the profiles for these accessions are lacking medium multiplex scores in the combined dataset (Venison and Ordidge 2022).
Out of the 1987 profiles that were produced for trees considered to be correct, 259 (13%) were found to have discrepancies from the profiles reported in the original data (Fernandez-Fernandez 2010) (Online Resource 1). One hundred and ninety-seven of these (10% of the total) were discrepancies in allele calling but all were deemed to be a matter of differential scoring rather than sample error. Of these, 104 cases (53%) represented instances where the original profile was deemed to be either more complete or accurate, 66 cases (34%) represented instances where the reproduction of the profile justified a change in allele size calling from the original call and 27 cases (14%) represented instances where additional alleles were added. A complete change in the data entry was deemed to be necessary in 62 cases (3% of the total) and this was because either the correct profile was missing (due to samples being missed or due to sampling error resulting in false double entries) or because two samples had been inadvertently swapped around in the prior analysis. Of these complete errors, 22 (35%) had been previously identified on the basis of having reported false duplication in the prior dataset and 26 (42%) were completely unexpected, the remaining 14 (23%) were simply missing entries. Fourty-one further entries were entirely new as these were for trees that had been accessed after the time of the prior analysis. A small number of adjustments to allele calling were made to the non-reproduced profiles from prior analysis on the basis of comparison to the reanalysis of samples deemed to be clonal replicates.
Identification of novel material in local collections
In the analysis of material submitted by the local collections a total of 3373 profiles were produced during 2016–2019. Of these, 2112 (63%) were judged to match with existing accessions in the NFC (including nine examples that were later deemed to match following MUNQ allocation) and 1261 (37%) were judged not to be present in the NFC. Of the profiles not represented in the NFC: 71 samples (6%) were judged to match another sample identified previously by the fingerprinting of other reference collections (mostly in Ireland); 494 samples (39%) were grouped as being indistinguishable from at least one other sample within the scheme, and 696 samples (55%) were distinct from any other sample (Online Resource 3).
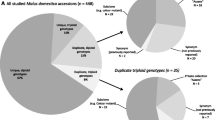
Of the 2112 samples identified to match accessions in the NFC, approximately 560 cultivars were represented (accepting that in a few cases the cultivar identification was not completely resolved due to either missing data or uncertain allele calls); the most common cultivar to be identified was ‘Bramley’s Seedling’ followed by ‘Blenheim Orange’ and ‘Newton Wonder’ (Fig. 1). The 494 samples distinguishable from the contents of the NFC and other reference collections but replicated within the scheme, were judged to represent 176 duplicate groups by MUNQ code. In agreement with minor allele calling discrepancies and occasional missing data, the groupings included nine MUNQ subcodes (Online Resource 2). Members of these duplicate groups were often distributed across multiple sites; the most common were identified on between seven and 13 occasions from six to eight sites (Online Resource 3). Overall, 127 of the duplicate groups distinguishable from the NFC contained samples identified across two or more sites.
Genetic diversity in the local collections
Allele number across both sets of samples ranged from 21 to 38 with an average of 30.7 alleles per locus (Table 1). The NFC and fruitID scheme samples had similar mean allele numbers (24.7 and 25.7, respectively) although there was a clear difference in the individual allele number for some loci between the two sample sets (specifically CH01h01 and CH04e05 where allele number differed by 14 and nine, respectively). The mean numbers of rare (frequency < 0.05) and private (present in only one sample set) alleles were also similar in the NFC and fruitID scheme samples (averaging 19 and 19.7, and 5 and 6, respectively); however, there remained a marked difference in the rare and private allele numbers for CH01h01 and CH04e05 (differing by 13 and nine alleles in the rare and 14 and nine alleles in the private, respectively). As a consequence, the range in the number of private alleles was larger in the fruitID scheme samples (one to 17 as opposed to one to seven in the NFC samples). All private alleles were classified as rare and consequently the average number (24.8) and range (15 to 34) of rare alleles across the whole dataset was larger than in each of the sample sets. The allele size range differed between the NFC and fruitID scheme samples due to the presence of private alleles at the extremes of the range, although this was most noticeable for CH01f03b and CH04e05 (where the size range differed by 39 bp and 22 bp, respectively). However, for the common alleles (freq > 0.05) the allele number from both the NFC and fruitID scheme samples was highly similar (differing by a maximum of two alleles for GD12 and either zero or one for all other loci) and was also highly similar to that of the combined dataset (differing by a maximum of one allele per locus).
Gene diversity (Nei 1978) was similar in the NFC and fruitID scheme samples and ranged from 0.654 to 0.902 and 0.665 to 0.910 with an average across all loci of 0.800 and 0.812, respectively. The combined non-exclusion probability, generated only from the NFC samples, ranged from 5.64 × 10–6 when including full-sibs to 1.06 × 10–15 when full-sibs were not considered. The combined non-exclusion probability when considering only the eight locus set ranged from 3.18 × 10–4 (full sibs) to 9.75 × 10–11 (unrelated).
Initial analysis of principal coordinates found a small number of entries (16 NFC, 9 fruitID) to appear as clear outliers. On checking, these were all missing substantial amounts of data: the 16 NFC samples represented all of those that were missing data for all four loci in the medium multiplex scores; the fruitID scheme samples were missing data for between one and six loci. In all cases samples were missing data for GD12. These 25 samples were removed and re-analysis of the remaining 2792 samples by principal coordinates analysis (Fig. 2) found that the two sets of samples are not differentiated and that little variation was explained by the main two coordinates (3.62 and 3.11%, respectively). This was supported by the analysis of molecular variance carried out both before and after the removal of samples which revealed that 99% of the molecular variance was found within the sample sets, with only 1% of the variance explained by differences among the two sample sets. In line with this, the FST values generated across all loci and the full set of (2817) samples indicated that genetic differentiation between the NFC and fruitID scheme samples was very low with values per locus ranging from FST = 0.002 to FST = 0.0054 with a mean of FST = 0.0035.
Discussion
Demonstration of use of SSRs in collection management
Our findings provide a clear demonstration of the utilisation of SSR marker technology in the active management and repropagation of germplasm collections. As far as we are aware, despite the large number of analytical studies that have created reference datasets, this is the first reported case of the use of genetic markers to support wholesale repropagation in a germplasm collection of apple.
In addition to the management of germplasm, the necessity to repeat the analysis of accessions served to test and improve the overall standard of the prior dataset and 92 profiles were improved through usage. A further 62 additional profiles were produced for samples that had been either erroneously sampled or handled in the prior analysis. It is notable that in the vast majority of cited examples of SSR analysis within germplasm collections, with the exception of Testolin et al. (2019), no thorough replication of sample analysis was reported to have been carried out. Subsets of samples were systematically replicated by Patzak et al. (2012) and Urrestarazu et al. (2016) for the purposes of checking reproducibility or aligning pre-existing data, but it appears to be the norm for most genetic analyses of fruit tree germplasm collections to be unreplicated. We acknowledge that this is a practicable solution to what is often a limitation in funding for such work. In line with this, the prior analysis of the NFC collection itself was not, with the exception of a few problematic profiles, replicated until now (Fernandez-Fernandez 2010) and neither was the genetic analysis for the purposes of identification within the fruitID scheme samples that we report here. The percentage of wholly corrected profiles was relatively low (3%), reflecting only a small number of sampling errors, and we would expect that most general analyses of diversity would not be greatly affected by this. However, instances of error would confound efforts to identify new material of matching genotypes and there will no doubt be cases of sampling error that become apparent in any attempts to align datasets across multiple countries (as has already been found to be the case during MUNQ allocation). Previous authors have noted that sampling or experimental error remains a possible explanation for a small number of otherwise unexplainable findings in such analyses (Ordidge et al. 2021; van Treuren et al. 2010).
One important sampling error from the prior data that was resolved in this analysis related to an original switch in sampling between the accessions 1926–023 Saltcote Pippin and 1955–008 Rubens (itself notable as a prior use of the name to that of the modern cultivar ‘Civni’). The consequence of this original error was that the allocation of the accession name to MUNQ code was originally incorrect and therefore the finding in Muranty et al. (2020) that MUNQ 767 was an offspring of ‘Knobby Russet’ and ‘Cox’s Orange Pippin’ (MUNQs 1946 and 163 respectively) was, in fact, a finding for ‘Saltcote Pippin’ as the corrected accession name for MUNQ 767. This corrected relationship is in agreement with the updated SSR dataset and dates of origin for the cultivars (Smith 1971).
Number of loci required for identification
We determined that the subset of eight loci was sufficient to distinguish practically all apart from the clonal relatives in the collection, the likely single exception being a pair of full siblings and we took this to confirm that they were sufficient for the identification of repropagated material. Of the 14 cases in which profiles were indistinguishable using eight loci but distinguishable by 12 loci and the additional profiles were only differentiated by either null alleles or 1–2 bp size differences, all the accessions had previously been placed within the same MUNQ group as those that they matched by eight loci (Denancé et al. 2020); and in the 12 of these groupings that had been analysed by DArT, all were previously considered to be indistinguishable (Ordidge et al. 2018). The instance in which accessions held under cultivar names believed to be synonymous (2011–042 Reinette du Mans and 1947–143 De Jaune) were identical across eight, but not across 12 loci was judged to most likely reveal a potential error in the data, and this would be worthy of further investigation. The instance in which a pair of accessions of cultivars documented to be full siblings (1948–022 Downton Pippin and 2000–102 Yellow Ingestrie) could not be distinguished using the eight loci set may indicate a genuine limit in the ability to distinguish by eight loci alone. The former of these last two cases had already been given MUNQ subcodes in recognition of this uncertainty and the latter had simply been given different MUNQ codes as recognition of them being truly independent genotypes (Denancé et al. 2020). Of the 16 instances in which accessions with differing genotypes for the eight loci set had been allocated to the same MUNQ group (again, where the differentiating factors were all either null alleles or allele size differences of 1–2 bp), 15 had been analysed by DArT and all of these groupings had been previously considered to be indistinguishable (Ordidge et al. 2018) in agreement with their MUNQ allocation. It is possible that these represent allele calls that could still be further refined and an equivalent level of minor variation was accepted in the allocation of MUNQ groupings (Denance et al., in prep). It is also notable that in a small number of cases, we reproduced allele scores which were not in complete agreement with clonal groupings (e.g. 1973–092 Golden Morspur, a documented sport of Golden Delicious that is differentiated by a missing [222 bp] allele in CH02d08 and an enlarged [219 vs 217 bp] allele in CH02c11 from all other Golden Delicious clones in the collection; and a variability in the presence/absence of the 118 bp allele of Hi02c07 in a group of documented Belle de Boskoop clones), or ploidy allocation (e.g. 2004–049 Queen Mary and 1975–317 Mother which consistently report a third allele [129 bp] for CH04c07 in an otherwise apparently diploid profile) and these may be worthy of further investigation.
With specific respect to the apparent CH02c11 allele shift in 1973–092 Golden Morspur, it was also notable that in the confirmation of profiles for accession 1947–051 Ijzerappel, an initial difference in one of the allele scores at this locus (again, 217 vs 219 bp) was observed from the prior dataset. However, on repeating the analysis from both of the original NFC 2 trees (on one of which the prior dataset was based) it was found that the two trees differed in the scoring of this allele, with one tree scoring 217 bp and the other scoring 219 bp and it would appear that the NFC3 replicates had both been grafted from tree 47_64 which reported the smaller allele despite the original trees being considered to be clonal.
All of the studies previously cited analysed between seven (Routson et al. 2009) and 21 (Lassois et al. 2016) SSR loci, with the exception of Evans et al. who analysed 80 loci, as part of their wider attempt to identify quantitative trait loci through pedigree-based analysis (Evans et al. 2011). In the case with fewest markers (Routson et al. 2009) the authors supposed that any matching profiles were an indication of material being clonally related and therefore samples representing either synonymous or clonally related cultivars, or having been mislabelled. A number of further studies deemed either eight or nine SSR loci to be sufficient for comparison of material and the assessment of basic genetic diversity (Garkava-Gustavsson et al. 2013; Gasi et al. 2016; Gharghani et al. 2009; Gross et al. 2014, 2012, 2018; Hokanson et al. 1998; Magby et al. 2019). The non-exclusion probabilities we report for the NFC dataset are of similar magnitude in comparison to the same set of loci studied by Lassois et al. who reported findings for individual loci that compute to 5.2 × 10–16 (12 locus set, unrelated) and 3.18 × 10–11 (eight locus set, unrelated) (Lassois et al. 2016) although are proportionally higher in comparison to a different set of nine loci studied by Gross et al. (2018) who reported a probability of 5.4 × 10–14 (unrelated). We would agree that this number of loci is sufficient for germplasm management, although a larger locus set allows for an element of redundancy in studies that are not replicated, such that occasional erroneous calls can be accepted or ignored. Studying larger numbers of loci also allows for a more detailed assessment of genetic diversity and structure (Lassois et al. 2016) although this was not the objective of our study, since both diversity and structure have been assessed in other studies using part, or all, of the NFC collection (Ordidge et al. 2018; Urrestarazu et al. 2016).
Value of the dataset
The dataset associated with this work has the potential to make a valuable contribution towards the efforts of the European Cooperative Program for plant Genetic Resources. We have incorporated the MUNQ values allocated to the NFC genotypes through the previous (Urrestarazu et al. 2016) and ongoing alignment of data (Denancé et al. 2020). The NFC is widely recognised, largely thanks to the work of previous curators, as one of the best-verified sources of true to type cultivar accessions in Europe and the collection has the potential to aid with resolving accession queries across numerous collections. We also expect, given the nature of distribution and historical renaming of many old cultivars of apple, that alignment with other international collections will further aid with the curation of the NFC collection itself.
Identification of wider diversity (with local value)
As is generally the case in apple, gene diversity scores for both sample sets were high and this is in keeping with apple being a self-incompatible outcrossing species. However, the overall measures of genetic diversity that we report failed to clearly distinguish between the fruitID scheme samples and the NFC. Principal coordinates analysis was unable to distinguish between the groupings and this was in line with a very low mean value for FST (0.0035 P < 0.0001) between the two sets. Much larger, but still relatively low values for FST have been reported for the analysis of local collections from Spain (0.057 ≤ FST≤ 0.12) (Pereira-Lorenzo et al. 2007; Pereira-Lorenzo et al. 2018; Pereira-Lorenzo et al. 2008; Pereira-Lorenzo et al. 2017), Bosnia and Herzegovina (FST = 0.06) (Gasi et al. 2010), Portugal (FST = 0.074) (Ferreira et al. 2016), and France (FST = 0.048 [old cider cultivars] FST = 0.031 [old dessert cultivars]) (Lassois et al. 2016) when compared against sets of more modern international and commercial cultivars and for the comparison of local material from different European regions (0.015 ≤ FST ≤ 0.042) (Urrestarazu et al. 2016). Differing results have been found from studies in Italy where FST values of 0.056 and 0.147 have been reported (Liang et al. 2015; Marconi et al. 2018), the higher value being specific to Central Italian local material and being generated after identifying groups by Bayesian analysis and removing the admixed accessions. A lower level of differentiation (FST = 0.014) was found between old dessert and old cider accessions in France (Lassois et al. 2016) and it is notable that we did not attempt to distinguish between the potential use of accessions in our analysis.
Whilst our use of FST to compare between material in local collections and an international genebank is slightly different to many of these previous studies, the primary objective of our analysis was to test whether the material identified through the fruitID scheme would clearly add to the genetic diversity maintained in the NFC collection. The lack of differentiation in our analysis may, in part, be a reflection of both the coverage of the NFC collection, and the range of material in local collections within the UK. The NFC collection has developed over many years (the earliest accessions dating to 1905). The objectives of the collection have ranged from acting as a reference source for named cultivars, through the collection of local and commercially interesting international cultivars in line with commercial trialling and the assessment of plant variety rights, to its current primary objective as a genetic resource. It would seem that overall, on the basis of our analysis, the local collections included in the fruitID scheme do not clearly differentiate from the range of genetic diversity held within the collection, although a more detailed analysis of genetic diversity and potential subpopulation structure would be required to investigate this further. We would note that our previous findings on the NFC collection as a whole, were that analysis of structure revealed a “complex and historically admixed group for which clear clustering was challenging” (Ordidge et al. 2018). Only 9–10% of accessions were strongly allocated to any of the major clusters and no particularly clear geographical differentiation was identified. For example, there was not a distinct cluster of UK material equivalent to that reported from studies in Spain (Pereira-Lorenzo et al. 2017). A similar lack of distinct clustering was reported from analysis of local material in the Danish genebank (Larsen et al. 2017). Given the very low value we report for FST (0.0035 P < 0.0001) and the result of AMOVA, the finding by Urrestarazu et al. (2015) that the priority marker sets (of up to 12 SSR loci) are only able to detect major divisions or moderate (FST > 0.05) differentiation, would suggest that additional loci would need to be included to make such an analysis meaningful.
In a study with similarity to our own, relatively little differentiation was reported between a set of local material in Northern Minnesota and the USDA collection (Gross et al. 2018), which in many ways is equivalent to the NFC collection in already containing a large range of both local, commercial and international cultivars. Conversely, the lack of overall differentiation could be taken to suggest that the majority of cultivars assessed in the fruitID scheme are likely to originate from a similar range of material to that which has been included within the NFC collection, and this would reflect the fact that many cultivars of apple have been brought into the UK from Europe and the rest of the world over the past centuries. Whilst many of the fruitID scheme samples were cultivars found in old orchards and gardens, a number of them were feral seedlings and these findings are also in agreement with the finding that feral seedlings in southern Canada were mostly descendants from the cultivated gene pool (Cronin et al. 2020).
Of the samples identified within the fruitID scheme, it was perhaps not surprising that the most commonly identified cultivars are all well-known, the majority of which originate from the UK before the start of the twentieth century. It is also notable that four out of twelve of them are triploids.
Although overall genetic diversity was not readily distinguishable, a number of novel and potentially rare alleles were identified in the fruitID scheme samples and it is possible that these reflect elements of diversity that are not currently represented in the NFC. As with all unreplicated analysis, this finding should perhaps be treated with an element of caution, and in a number of cases the private alleles were scored at single integers in an otherwise di-nucleotide repeat; a thorough check on allele calling for loci exhibiting large numbers of rare alleles offset from the more common allele range would be valuable to confirm whether these are reproducible. Perhaps with the addition of further loci, it could also be possible to investigate these alleles through the identification of potential parent–offspring relationships, in which incorrectly allocated alleles would appear as inconsistent Mendelian errors.
Nonetheless, many of the trees that were found to be unmatched in the NFC have been identified as having local value, sufficient for them to be included within private and non-governmental collections and some of them are proposed as potential findings of older cultivars. All of them were deemed to be sufficiently interesting to commission the generation of a DNA fingerprint, either in an attempt to identify them, or in order to confirm their uniqueness (a number of the samples were presumed or known seedlings). Whilst there is currently no plan to expand the national system to include decentralized holdings, it would be worthy to note that there remain genetically distinguishable accessions of material that have been deemed to have sufficient local and cultural value to be maintained by the local collections outside of the NFC. A number of genotypes were found repeatedly within the fruitID scheme but could not be identified against the NFC database. That they were found in multiple places is an indication that they have been put into cultivation and distributed by grafting and again, this can be taken as an indication that they have previously been identified to have value.
In an attempt to address the large number of novel cultivars and seedlings that had been identified through the fruitID scheme, a UK Register of Local Cultivars has been launched. Like the fruitID scheme itself, this has largely been a community-led initiative and a panel of experts from around the UK has been assembled in order to agree a consensus position on the naming of any material found to be genetically distinguishable (fruitID 2021). The scheme requires applicants to present a case for naming of any material reporting a profile that is not represented in the NFC (or a case to challenge current NFC accession naming where it is felt that a more correct cultivar name has been identified). At the time of writing, 57 of the duplicate groups and 134 of the unmatched samples had received a naming accreditation to be accepted by both the NFC and the UK local groups. Highlights of the scheme so far have been the acceptance of credible candidates for the old cultivars ‘Barcelona Pearmain’ (dating to 1831 and previously thought to be lost) and ‘Bringewood Pippin’ (dating to 1800 and also previously thought to be lost), the naming of a number of new seedling cultivars Including ‘Don’s Delight’ (A081) ‘Forty Winks’ (A927) and ‘White Tie’ (A2248) and the identification of potentially valuable sports of known cultivars such as ‘Blofield Russet’ (A4340) as a full russet sport of the cultivar ‘Greensleeves’. The scheme has also led to the potential identification and renaming of a small number of NFC accessions whose identity was either unknown or in doubt, with particular examples being a correction to the identification of accession 1991–019 (formerly Hereford White) identified to be ‘Golden Ball’ and the identification of a potentially historical mix-up in the NFC accessions of ‘Hunt’s Duke of Gloucester’ (1942–002, formerly labelled Puckrupt Pippin) and the NFC accession of ‘Puckrupp Pippin’ (1948–508, formerly labelled Hunt’s Duke of Gloucester). In all cases, it is possible that the currently accredited UK name is further amended following the comparison of material in other countries.
Opportunity for community engagement
The relative accessibility of SSR data, requiring little bioinformatic support, offers an opportunity to bridge the gap between germplasm collections such as the NFC and both local collections and collectors. At the time of writing, the scheme had included 168 participants ranging from institutions and NGOs managing local collections through to private individuals with a single unknown tree. No doubt, this opportunity will be more relevant to the range of crops that are likely to be maintained in parallel by the volunteer community, of which apple is perhaps the prime example. The fruitID DNA scheme data are tracked in real-time by a compilation sheet of all samples submitted and their outcomes and cross references. The scheme also maintains a spreadsheet of genotypes in the NFC, and other reference collections alongside the non-matching scheme sample results. This provides rapid search and comparison for the user community. It also has a method of finding plausible parentage where the data exists within the dataset (fruitID 2021).
Furthermore, as an unplanned consequence, it was possible to identify profiles in the fruitID dataset that were deemed to match samples that were lacking data (specifically where the medium multiplex had either failed or been missed) in the NFC dataset. Scores have been imputed from samples A568, A1277 and A2642 to NFC accessions 1974–347 Grenadier, 1950–041 Sandew and 1945–101 Green Purnell (Online Resource 2; Venison and Ordidge 2022) although these imputed scores were not used in the analysis of genetic diversity. All of these were supported by MUNQ allocation and the latter was identified within a group that also included trees originally sourced from the NFC accessions for the purposes of morphological comparison.
Relevance of SSR and need for equivalence in other approaches
Recent authors (Testolin et al. 2019) discuss the potential added value that tri-nucleotide repeats might add to the ease of scoring, consistency of data and consequently the ability to compare data between labs, and this approach has also been discussed and proposed in both blueberry (Bassil et al. 2020) and pear (Zurn et al. 2020). However, despite ongoing concerns about the possibility of aligning SSR data between labs, significant efforts have been made to align data through both EU-funded (Urrestarazu et al. 2016) and ECPGR-supported (Denancé et al., in prep; Ordidge et al. 2021) projects. Datasets, including the ECPGR locus set, are currently being compiled. Additionally, potential efficiency improvements to the use of the ECPGR locus set have been proposed (Cmejlova et al. 2021).
Alternative marker technologies are available and have been proven to be useful in studies around both the identification of individuals and pedigrees (Harper et al. 2020; Howard et al. 2017; Vanderzande et al. 2017; Winfield et al. 2020). A number of these newer technologies would seem to potentially offer an increase in efficiency, at least at the level of data generation. The value of the established datasets using SSR technology should however, not be underestimated in any considerations by the research community to move towards different technologies; it would seem necessary to consider how the established database would be recreated in any move to an otherwise incompatible marker type. Nonetheless, co-ordination in any technology will likely be less than perfect and there remain a number of recent SSR-based studies that have used novel sets of loci which will not be able to be aligned on an international basis.
Conclusions
We would conclude that SSR marker technology is a useful tool for both the management of germplasm, and the comparison of locally held material to germplasm collections. The use of such technology in the active management of repropagation allows the data to be tested, and consequently either improved or verified in cases where replication is not possible in the first instance. We would also note that there clearly remain genetically distinguishable accessions in the UK which are valued locally and offer the potential to complement national holdings of germplasm, although establishing the true genetic value of this material would require further, more detailed analysis.
Availability of data and material
All data used in the publication are available in the Online Resources or cited datasets. Material from the NFC is available upon request through the corresponding author.
Code availability
Not applicable.
References
Bassil N, Bidani A, Nyberg A, Hummer K, Rowland LJ (2020) Microsatellite markers confirm identity of blueberry (Vaccinium spp.) plants in the USDA-ARS National Clonal Germplasm Repository collection Genetic Resources and Crop Evolution. https://doi.org/10.1007/s10722-019-00873-8
Bramel PJ, Volk G (2019) A global strategy for the conservation and use of apple genetic resources. Global Crop Divers Trust Bonn Germany. https://doi.org/10.13140/RG.2.2.34072.34562
Cabe PR, Baumgarten A, Onan K, Luby JL, Bedford DS (2005) Using microsatellite analysis to verify breeding records: A study of “Honeycrisp” and other cold-hardy apple cultivars. HortScience 40:15–17. https://doi.org/10.21273/hortsci.40.1.15
Cmejlova J, Rejlova M, Paprstein F, Cmejla R (2021) A new one-tube reaction kit for the SSR genotyping of apple (Malus × domestica Borkh). Plant Sci. https://doi.org/10.1016/j.plantsci.2020.110768
Cronin D, Kron P, Husband BC (2020) The origins and evolutionary history of feral apples in southern Canada. Mol Ecol 29:1776–1790. https://doi.org/10.1111/mec.15277
Denancé C, Muranty H, Durel C-E (2020) MUNQ—Malus UNiQue genotype code for grouping apple accessions corresponding to a unique genotypic profile, V1 edn. Portail Data INRAE. https://doi.org/10.15454/HKGMAS
Edge-Garza DA, Rowland TV, Haendiges S, Peace C (2014) A high-throughput and cost-efficient DNA extraction protocol for the tree fruit crops of apple, sweet cherry, and peach relying on silica beads during tissue sampling. Mol Breed 34:2225–2228. https://doi.org/10.1007/s11032-014-0160-x
Evans KM et al (2011) Genotyping of pedigreed apple breeding material with a genome-covering set of SSRs: trueness-to-type of cultivars and their parentages. Mol Breed 28:535–547. https://doi.org/10.1007/s11032-010-9502-5
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes—application to human mitochondrial-DNA restriction data. Genetics 131:479–491
Fernandez-Fernandez F (2010) Fingerprinting the National apple and pear collections. Final report of the Defra research project GC0140. http://randd.defra.gov.uk/Document.aspx?Document=GC0140SID5FingerprintingFernandez.pdf. Accessed 20 July 2021
Fernandez-Fernandez F (2013) Common set of ECPGR SSR markers for Malus characterization. http://archive-ecpgr.cgiar.org/fileadmin/www.ecpgr.cgiar.org/NW_and_WG_UPLOADS/MalusPyrus2012/MalusPyrus_SSR_Markers.pdf. Accessed 21 July 2021
Ferreira V et al (2016) Genetic pool structure of local apple cultivars from portugal assessed by microsatellites. Tree Genet Genom 12:36. https://doi.org/10.1007/s11295-016-0997-8
Flachowsky H HM (2011) The network of the 'German National Fruit Genebank', a new concept for sustainable preservation of fruit genetic resources. http://archive-ecpgr.cgiar.org/fileadmin/www.ecpgr.cgiar.org/NW_and_WG_UPLOADS/Prunus/German%20National%20Fruit%20Genebank.pdf. Accessed 20 July 2021
fruitID (2021). https://www.fruitid.com/#help. Accessed 18 Jan 2021
Garkava-Gustavsson L, Brantestam AK, Sehic J, Nybom H (2008) Molecular characterisation of indigenous Swedish apple cultivars based on SSR and S-allele analysis. Hereditas 145:99–112. https://doi.org/10.1111/j.2008.0018-0661.02042.x
Garkava-Gustavsson L, Mujaju C, Sehic J, Zborowska A, Backes GM, Hietaranta T, Antonius K (2013) Genetic diversity in Swedish and Finnish heirloom apple cultivars revealed with SSR markers. Sci Hortic 162:43–48. https://doi.org/10.1016/j.scienta.2013.07.040
Gasi F et al (2016) Redundancies and genetic structure among ex situ apple collections in Norway examined with microsatellite markers. HortScience 51:1458. https://doi.org/10.21273/hortsci11212-16
Gasi F, Simon S, Pojskic N, Kurtovic M, Pejic I (2010) Genetic assessment of apple germplasm in Bosnia and Herzegovina using microsatellite and morphologic markers. Sci Hortic 126:164–171. https://doi.org/10.1016/j.scienta.2010.07.002
Gharghani A et al (2009) Genetic identity and relationships of Iranian apple (Malus x domestica Borkh.) cultivars and landraces, wild Malus species and representative old apple cultivars based on simple sequence repeat (SSR) marker analysis. Genetic Resour Crop Evol 56:829–842. https://doi.org/10.1007/s10722-008-9404-0
Gross BL, Henk AD, Richards CM, Fazio G, Volk GM (2014) Genetic diversity in Malus x domestica (Rosaceae) through time in response to domestication. Am J Bot 101:1770–1779. https://doi.org/10.3732/ajb.1400297
Gross BL, Volk GM, Richards CM, Forsline PL, Fazio G, Chao CT (2012) Identification of duplicate accessions within the USDA-ARS national plant germplasm system malus collection. J Am Soc Horticult Sci 137:333–342. https://doi.org/10.21273/jashs.137.5.333
Gross BL, Wedger MJ, Martinez M, Volk GM, Hale C (2018) Identification of unknown apple (Malus × domestica) cultivars demonstrates the impact of local breeding program on cultivar diversity. Genet Resour Crop Evol 65:1317–1327. https://doi.org/10.1007/s10722-018-0625-6
Guarino C, Santoro S, De Simone L, Lain O, Cipriani G, Testolin R (2006) Genetic diversity in a collection of ancient cultivars of apple (Malus x domestica Borkh.) as revealed by SSR-based fingerprinting. J Horticult Sci Biotechnol 81:39–44
Guilford P, Prakash S, Zhu JM, Rikkerink E, Gardiner S, Bassett H, Forster R (1997) Microsatellites in Malus X domestica (apple): abundance, polymorphism and cultivar identification. Theor Appl Genet 94:249–254. https://doi.org/10.1007/s001220050407
Hardy OJ, Vekemans X (2002) SPAGEDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes 2:618–620. https://doi.org/10.1046/j.1471-8286.2002.00305.x
Harper H et al (2020) The Long Ashton Aegacy: Characterising United Kingdom West Country cider apples using a genotyping by targeted sequencing approach. Plants People Planet 2:167–175. https://doi.org/10.1002/ppp3.10074
Hogg R (1884) The Fruit Manual. Fifth Edition. Journal of Horticultural Office, London.
Hokanson SC, Szewc-McFadden AK, Lamboy WF, McFerson JR (1998) Microsatellite (SSR) markers reveal genetic identities, genetic diversity and relationships in a Malus x domestica borkh. core subset collection. Theor Appl Genet 97:671–683. https://doi.org/10.1007/s001220050943
Howard NP et al (2017) Elucidation of the ‘Honeycrisp’ pedigree through haplotype analysis with a multi-family integrated SNP linkage map and a large apple (Malus×domestica) pedigree-connected SNP data set. Horticult Res 4:17003. https://doi.org/10.1038/hortres.2017.3
Irish BM, Cuevas HE, Simpson SA, Scheffler BE, Sardos J, Ploetz R, Goenaga R (2014) Musa spp. Germplasm management: microsatellite fingerprinting of USDA-ARS National Plant Germplasm System collection. Crop Sci 54:2140–2151. https://doi.org/10.2135/cropsci2014.02.0101
Jamshidi S, Jamshidi S (2011) NTSYSpc 2.02 e implementation in molecular biodata analysis (clustering, screening, and individual selection). In: 4th International conference on environmental and computer science (ICECS 2011). Singapore, 16–18 September. IACSIT Press.
Kalinowski ST, Taper ML , Marshall TC (2007) Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol Ecol 16:1099–1106. https://doi.org/10.1111/j.1365-294x.2007.03089.x
Kellerhals M, Egger S Inventory on fruit genetic resources in Switzerland. In: 2004 International society for horticultural science (ISHS), Leuven, Belgium, pp 585–588. https://doi.org/10.17660/ActaHortic.2004.663.102
Kim JH et al (2019) Genetic diversity structure, and core collection of Korean apple germplasm using simple sequence repeat markers. Horticult J 88:329–337. https://doi.org/10.2503/hortj.UTD-041
Kitahara K, Matsumoto S, Yamamoto T, Soejima J, Kimura T, Komatsu H, Abe K (2005) Parent identification of eight apple cultivars by S-RNase analysis and simple sequence repeat markers. HortScience 40:314–317. https://doi.org/10.21273/hortsci.40.2.314
Larsen B, Toldam-Andersen TB, Pedersen C, Orgaard M (2017) Unravelling genetic diversity and cultivar parentage in the Danish apple gene bank collection. Tree Genet Genomes. https://doi.org/10.1007/s11295-016-1087-7
Lassois L et al (2016) Genetic diversity, population structure, parentage analysis, and construction of core collections in the French apple germplasm based on SSR markers. Plant Mol Biol Report 34:827–844. https://doi.org/10.1007/s11105-015-0966-7
Liang W, Dondini L, De Franceschi P, Paris R, Sansavini S, Tartarini S (2015) Genetic diversity, population structure and construction of a core collection of apple cultivars from Italian germplasm. Plant Mol Biol Report 33:458–473. https://doi.org/10.1007/s11105-014-0754-9
Magby J, Volk GM, Henk A, Miller S (2019) Identification of historic homestead and orchard apple cultivars in Wyoming. HortScience 54:8–16. https://doi.org/10.21273/hortsci13436-18
Marconi G, Ferradini N, Russi L, Concezzi L, Veronesi F, Albertini E (2018) Genetic characterization of the apple germplasm collection in central Italy: the value of local varieties. Front Plant Sci. https://doi.org/10.3389/fpls.2018.01460
Moriya S, Iwanami H, Okada K, Yamamoto T, Abe K (2011) A practical method for apple cultivar identification and parent-offspring analysis using simple sequence repeat markers. Euphytica 177:135–150. https://doi.org/10.1007/s10681-010-0295-8
Muranty H et al (2020) Using whole-genome SNP data to reconstruct a large multi-generation pedigree in apple germplasm. BMC Plant Biol 20:2. https://doi.org/10.1186/s12870-019-2171-6
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590
Olasupo FO et al (2018) Genetic identity and diversity of Nigerian cacao genebank collections verified by single nucleotide polymorphisms (SNPs): a guide to field genebank management and utilization. Tree Genet Genom 14:16. https://doi.org/10.1007/s11295-018-1244-2
Ordidge M, Kirdwichai P, Baksh MF, Venison EP, Gibbings JG, Dunwell JM (2018) Genetic analysis of a major international collection of cultivated apple varieties reveals previously unknown historic heteroploid and inbred relationships. PLoS ONE 13:26. https://doi.org/10.1371/journal.pone.0202405
Ordidge M et al (2021) Towards a joint international database: alignment of SSR marker data for European collections of cherry germplasm. Plants 10:1243. https://doi.org/10.3390/plants10061243
Ordidge M, Venison E (2018) Simple Sequence Repeat (SSR) Fingerprint data for apples from the National Fruit Collection as used in Ordidge et al PLOS One 2018. University of Reading Research Data Archive. https://doi.org/10.17864/1947.163
Patzak J, Paprstein F, Henychova A, Sedlak J (2012) Genetic diversity of Czech apple cultivars inferred from microsatellite markers analysis. Hortic Sci 39:149–157
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295. https://doi.org/10.1111/j.1471-8286.2005.01155.x
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28:2537–2539. https://doi.org/10.1093/bioinformatics/bts460
Pereira-Lorenzo S, Ramos-Cabrer AM, Díaz-Hernández MB (2007) Evaluation of genetic identity and variation of local apple cultivars (Malus × domestica Borkh.) from Spain using microsatellite markers. Genet Resour Crop Evol 54:405–420. https://doi.org/10.1007/s10722-006-0003-7
Pereira-Lorenzo S et al (2018) Genetic diversity and core collection of Malus × domestica in northwestern Spain, Portugal and the Canary Islands by SSRs. Sci Hortic 240:49–56. https://doi.org/10.1016/j.scienta.2018.05.053
Pereira-Lorenzo S, Ramos-Cabrer AM, Gonzalez-Diaz AJ, Diaz-Hernandez MB (2008) Genetic assessment of local apple cultivars from La Palma, Spain, using simple sequence repeats (SSRs). Sci Hortic 117:160–166. https://doi.org/10.1016/j.scienta.2008.03.033
Pereira-Lorenzo S et al (2017) Analysis of the genetic diversity and structure of the Spanish apple genetic resources suggests the existence of an Iberian genepool. Ann Appl Biol 171:424–440. https://doi.org/10.1111/aab.12385
Routson KJ, Reilley AA, Henk AD, Volk GM (2009) Identification of historic apple trees in the Southwestern United States and implications for conservation. HortScience 44:589–594. https://doi.org/10.21273/hortsci.44.3.589
Salvi S, Micheletti D, Magnago P, Fontanari M, Viola R, Pindo M, Velasco R (2014) One-step reconstruction of multi-generation pedigree networks in apple (Malus x domestica Borkh.) and the parentage of Golden Delicious. Mol Breed 34:511–524. https://doi.org/10.1007/s11032-014-0054-y
Smith MWG (1971) National apple register of the United Kingdom. Ministry of Agriculture, Fisheries and Food, London.
Testolin R et al (2019) Genotyping apple (Malus x domestica Borkh.) heirloom germplasm collected and maintained by the Regional Administration of Friuli Venezia Giulia (Italy). Sci Hortic 252:229–237. https://doi.org/10.1016/j.scienta.2019.03.062
Urrestarazu J et al (2016) Analysis of the genetic diversity and structure across a wide range of germplasm reveals prominent gene flow in apple at the European level. BMC Plant Biol. https://doi.org/10.1186/s12870-016-0818-0
Urrestarazu J, Royo JB, Santesteban LG, Miranda C (2015) Evaluating the Influence of the microsatellite marker set on the genetic structure inferred in Pyrus communis L. PLoS ONE 10:e0138417. https://doi.org/10.1371/journal.pone.0138417
van Treuren R, Kemp H, Ernsting G, Jongejans B, Houtman H, Visser L (2010) Microsatellite genotyping of apple (Malus × domestica Borkh.) genetic resources in the Netherlands: application in collection management and variety identification. Genet Resour Crop Evol 57:853–865. https://doi.org/10.1007/s10722-009-9525-0
Vanderzande S, Micheletti D, Troggio M, Davey MW, Keulemans J (2017) Genetic diversity, population structure, and linkage disequilibrium of elite and local apple accessions from Belgium using the IRSC array. Tree Genet Genom 13:125. https://doi.org/10.1007/s11295-017-1206-0
Venison E, Ordidge M (2022) Simple Sequence Repeat (SSR) fingerprint data for apples from the National Fruit Collection as used in the article ‘Microsatellite markers as a tool for active germplasm management and bridging the gap between national and local collections of apple’, University of Reading Research Data Archive. https://doi.org/10.17864/1947.000351
Volk GM, McCardle K, Serin J, Magby J, Mazur R (2021) Genetic fingerprinting identifies apple (Malus domestica Borkh.) fruit cultivars in historic orchards on public lands. Genet Resour Crop Evol 1:2. https://doi.org/10.1007/s10722-021-01175-8
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population-structure. Evolution 38:1358–1370. https://doi.org/10.2307/2408641
Winfield M et al (2020) Development of a minimal KASP marker panel for distinguishing genotypes in apple collections. PLoS ONE 15:e0242940. https://doi.org/10.1371/journal.pone.0242940
Zurn JD, Nyberg A, Montanari S, Postman J, Neale D, Bassil N (2020) A new SSR fingerprinting set and its comparison to existing SSR- and SNP-based genotyping platforms to manage pyrus germplasm resources. Tree Genet Genom 16:72. https://doi.org/10.1007/s11295-020-01467-7
Acknowledgements
We would like to thank Prof. Raffaele Testolin, University of Udine and Dr Fuad Gasi, University of Sarajevo for their helpful advice on the analysis of genetic data. We would like to thank Maria Carcamo for her work on the early phases of analysis of fruitID scheme samples. We would like to thank all of the participants of the fruitID scheme for their submission of samples, agreement to make data available and for their work in identifying and conserving local cultivars of apple.
Funding
Support for this work was supplied by Defra (project code GC0147 The Long Term Housing, Maintenance and Scientific Curation of the [UK] National Fruit Collections). Support for the Fruit ID scheme was supplied by the UK local orchard groups.
Author information
Authors and Affiliations
Contributions
EV and SL carried out molecular genetic analysis and curated data for the NFC and fruitID scheme, respectively. CD and C-ED curated data for MUNQ allocation. FF-F curated data for the fruitID scheme. PL and MO conceptualized the experiment and curated data. PL administrated the fruitID scheme. MO carried out data analysis and wrote the first draft of the manuscript. EV, SL, CD, C-ED, FF-F and PL reviewed and edited the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
fruitID scheme data were generated through a commercial fingerprinting service offered by NIAB EMR, the organization for which SL and FF-F are employed. No other authors have interests to declare.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
All coauthors have read and agreed to submit the manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Venison, E.P., Litthauer, S., Laws, P. et al. Microsatellite markers as a tool for active germplasm management and bridging the gap between national and local collections of apple. Genet Resour Crop Evol 69, 1817–1832 (2022). https://doi.org/10.1007/s10722-022-01342-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-022-01342-5