Abstract
Whilst it is established that almost all cultivated coffee (Coffea arabica L.) varieties originated in Yemen after some coffee seeds were introduced into Yemen from neighboring Ethiopia, the actual coffee genetic diversity in Yemen and its significance to the coffee world had never been explored. We observed five genetic clusters. The first cluster, which we named the Ethiopian-Only (EO) cluster, was made up exclusively of the Ethiopian accessions. This cluster was clearly separated from the Yemen and cultivated varieties clusters, hence confirming the genetic distance between wild Ethiopian accessions and coffee cultivated varieties around the world. The second cluster, which we named the SL-17 cluster, was a small cluster of cultivated worldwide varieties and included no Yemen samples. Two other clusters were made up of worldwide varieties and Yemen samples. We named these the Yemen Typica-Bourbon cluster and the Yemen SL-34 cluster. Finally, we observed one cluster that was unique to Yemen and was not related to any known cultivated varieties and not even to any known Ethiopian accession: we name this cluster the New-Yemen cluster. We discuss the consequences of these findings and their potential to pave the way for further comprehensive genetic improvement projects for the identification of major resilience/adaptation and cup quality genes that have been shaped through the domestication process of C. arabica.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
12.5 million households around the world receive an income from coffee growing (Browning 2018). Coffee is mainly produced by two species:: Coffea arabica L. producing Arabica coffee and Coffea canephora Pierre ex A.Froehner producing the coffee known as Conilon when produced in Brazil, and Robusta anywhere else in the world. Arabica coffee production is facing multiple challenges such as climate change (Bunn et al. 2015) and serious diseases such as coffee leaf rust caused by Hemileia vastatrix (Avelino et al. 2015), and Coffee Berry Disease caused by Colletotrichum kahawae (Van der Vossen et al. 2015). Whilst there is no single solution to such complex challenges, the study of genetics and the breeding of improved varieties is a critical area of investigation for potential solutions. The search for genetically superior quality coffee varieties has become a central issue for the specialty market (Montagnon et al. 2019). Unfortunately, the genetic diversity of C. arabica is among the lowest in the cultivated crop, due to a recent single event of polyploidization (Scalabrin et al. 2020). Furthermore, the major part of this low genetic diversity is found mainly in Ethiopia and to a lesser degree in South Sudan (Chevalier 1929; Harlan 1969; Sylvain 1958; Thomas 1942).
Whilst Ethiopia and South Sudan are the center of origin of C. arabica, Yemen has been its key center of domestication. Both historical records (de la Roque 1716; Ukers 1922; Chevalier 1929; Cramer 1957; Haarer 1958; Meyer 1965; Koehler 2017) and past phenotypic (Montagnon and Bouharmont 1996) or genetic studies (Lashermes et al. 1996; Anthony et al. 2002; Silvestrini et al. 2007; Pruvot-Woehl et al. 2020; Scalabrin et al. 2020) indicate that all the Arabica varieties cultivated outside Ethiopia transited through the Yemen domestication center. Still, while all the previous genetic studies were using Yemeni coffee samples to check the Yemeni stop-over between Ethiopia and the spread of Arabica coffee varieties to the world, none has revealed the Yemeni genetic diversity. The few genetic studies focusing on Yemeni coffees (Al-Murish et al. 2013; Hussein et al. 2017) indicated the genetic heterogeneity of varieties as identified by Yemeni farmers. Neither Ethiopian accessions nor worldwide cultivated varieties were included in these studies.
Various molecular markers are available to geneticists and breeders for genetic studies and genome analysis, namely Single Nucleotide Polymorphism (SNPs) and Single Sequence Repeats (SSRs) (reviewed by Adhikari et al. 2017). Genotyping by Sequencing (GBS) technique which provides thousands of SNPs for a dense coverage of the genome is no doubt the ideal method to study the relationship between genotype and phenotype, namely in polyploids (Clevenger et al. 2015). SNP’s are often used in coffee for genome wide selection in C. canephora (Alkimim et al. 2020). Scalabrin et al. (2020) demonstrated the recent unique polyploidization event by sequencing the C. arabica genome using SNPs from the two sub-genomes of this tetraploid species. However, when the objective is fingerprinting or a genetic diversity study, the high level of polymorphism in SSRs renders it a reliable, practical and cost effective choice (Hodel et al. 2016). In grapes, the first approach to characterize a Vitis germplasm collections with ten SSRs proved a high discriminating capacity for grapevine varieties (Emmanuelli et al. 2013). In the same study, SSRs proved as efficient as SNPs to establish the genetic diversity of grapevine. Singh et al. (2013) found that SSR markers were more efficient than SNP markers when the objective was strictly the study of genetic diversity. Anthony et al. (2002) found that 6 SSR markers were efficient to confirm the origin of cultivated C. arabica varieties, spreading from Yemen after an early introduction from Ethiopia. More recently, da Silva et al. (2019) used 30 markers to efficiently discriminate between C. arabica varieties and three diploid Coffea species. Benti et al. (2020) used 14 SSR markers to efficiently discriminate between 40 cultivated C. arabica varieties in Ethiopia. Finally, Pruvot-Woehl et al. (2020) demonstrated that a set 8 SSR markers—used in the present study-used to genotype 2533 samples representing the largest known genetic diversity of C. arabica was efficient in discriminating between varieties and could be used for varietal authentication, and hence for genetic diversity studies.
Over the past half-decade, Qima Coffee (www.qimacoffee.com)—a coffee company working at the ground level in Yemen- has developed research activities in order to better understand the genetic landscape of Yemeni coffee. A breeding population made of 45 individuals representing various coffee morphotypes observed in Yemen has been gathered. Using this breeding population, together with Ethiopian accessions and cultivars as well as a representation of the cultivated varieties worldwide, we present here the first global study aiming at describing the C. arabica coffee genetic diversity in Yemen. The main questions addressed in the study are as follows: What is the magnitude and the structure of the genetic diversity in Yemen? How does the genetic diversity of Yemen compare with the known genetic diversity of coffee in Ethiopia and that of the cultivated C. arabica varieties worldwide? And finally, does Yemen’s genetic diversity offer opportunities for genetic improvement of C. arabica?
Material and methods
Plant material
137 samples of C. arabica were studied. They belong to the following three categories (Table 1).
Ethiopian accessions (EA)
72 accessions are representing the Ethiopian accessions collected in 1966 and 1968 by the FAO and Orstom survey, respectively (FAO 1968; Charrier 1978). Those 72 accessions were selected as they are part of the core collection defined by World Coffee Research and the Centro Agronómico Tropical de Investigación y Enseñanza (CATIE) in 2014 (Solano, personal communication).
Worldwide cultivars (WWC)
20 samples represent main cultivated varieties grown worldwide outside Ethiopia. This includes Bourbon and Typica, but also East African and Indian varieties that have been shown to have transited through Yemen from Ethiopia before being introduced in all present coffee producing countries (reviewed in Pruvot-Woehl et al. 2020).
Yemen Qima breeding population (YQ)
45 samples from the Qima breeding population, made up of 45 trees selected from Yemen germplasm representing the major coffee growing areas.
DNA extraction and SSR marker analysis
All the operation of DNA extraction and SSR marker analysis were performed by the ADNiD laboratory of the Qualtech company in the South of France (http://www.qualtech-groupe.com/en/).
Genomic DNA was extracted from approximately 20 mg of dried tissue according a homemade protocol with SDS buffer. DNA was then purified with magnetic bead (Agencourt AMPure XP, Beckman Coulter, Brea, California, USA) followed by elution in Tris Edta (TE) buffer.
The DNA concentration was estimated with a Enspire spectrofluorimeter (Perkin Elmer) with a bisbenzimide DNA intercalator (Hoechst 33,258) and by comparison with known standards of DNA.
Eight SSR primer pairs (Table 2) selected after Combes et al. (2000) and whose wide discrimination power was confirmed by Pruvot-Woehl et al. (2020) have been used.
PCR was performed in a 15 μL final volume comprising 30 ng genomic DNA and 7.5 μL of 2 × PCR buffer (Type-it Microsatellite PCR Kit, Qiagen), 1.0 μM each of forward and reverse primer (10 µM). Amplifications were carried out in thermal cycler (Eppendorf) programmed at 94 °C for 5 min for initial denaturation, followed by 94 °C for 30 s, annealing temperature depending on the primer used for 30 s and 72 °C for 1 min for 35 cycles followed by a final step of extension at 72 °C for 5 min. Final holding temperature was 4° C.
PCR samples were run on a capillary electrophoresis, ABI 3130XL with an internal standard: GeneScan 500 LIZ size standard (Applied Biosystems).
Alleles were scored using GeneMapper v.4.1 software (Applied Biosystems).
Data analysis
The method described by Pruvot-Woehl et al. (2020) was used. Because C. arabica is tetraploid, the presence/absence (1/0) was coded for each allele. Strictly speaking, we are dealing with SRR allelic phenotype rather than genotype. Indeed, the phenotype AB could be either of the following genotypes: AABB, ABAB, AAAB, ABBB.
DARwin6 software (Perrier and Jacquemoud-Collet 2006) was used with single data files. Dissimilarity matrix was calculated using Dice Index and was the basis for the construction of the genetic diversity tree using the weighted Neighbor-Joining method (Saitou and Nei 1987) and the execution of the Principal Coordinates Analysis (PCoA). As indicated by Perrier and Jacquemoun-Collet (2006), the PCoA give an overall representation of the diversity, while tree methods tend to represent individual genetic relationships faithfully. Hence, these two different ways of viewing the data are complementary.
In order to check the robustness of the clusters, a Discriminant Analysis (DA) (Tomassone et al. 1988) was run on the coordinates on the five first axis of the PCoA. The statistical difference between the genetic clusters was checked with the Wilks Lambda test. The percentage of good classification was checked through the cross-validation when each sample is classified based on the model build on the whole sample but this sample. The DA and related tests were performed with the Xlstat software (Addinsoft 2020).
Results
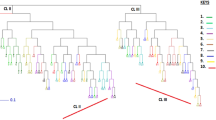
The neighbor-Joining tree based on all the samples (Fig. 1) shows five well marked different clusters. Sample categories are not evenly distributed in each cluster (Table 1). One major cluster was almost entirely comprised of Ethiopian accessions. We named it the Ethiopian Only cluster (EO). Five worldwide cultivated varieties belonged to that cluster: (i) Geisha (CATIE code T.02722) is the famous Geisha which originally became renowned in Panama (Pruvot-Woehl et al. 2020), (ii) Java is a variety originally selected in Cameroon (Bouharmont 1994) in an Ethiopian population, (iii) Chiroso is a name given to a variety grown in Colombia and said to have a superior cup quality (Montagnon Pers. Obs.), (iv) SL-06 was selected in Kenya in the early twentieth century, supposedly from a Kent tree (Jones 1956) and (v) Mibirizi is one of the first variety grown in the Great Lake region; its origin is unknown (Leplae 1936). The CATIE code of the Mibirizi accession in this cluster is T.02702.
The second cluster was made of Ethiopian accessions and worldwide cultivated varieties. Cultivated varieties were varieties selected in East Africa in the early twentieth century: SL-14, SL-17 and K-7. SL-14 and SL-17 were selected from a “Drought Resistant” population in Kenya. The origin of this “Drought Resistant” population is unknown (Jones 1956). Both K-7 and K-758 have the same genetic fingerprint and are supposed to descent from Kent selections (Fernie 1970). Mibirizi with CATIE Code T.03622 was also in that cluster. No Yemeni samples were found in that cluster. We named this cluster the “SL-17” cluster.
The third and fourth clusters were made only of worldwide coffee cultivars and Yemeni samples. One cluster included Yemeni accessions and two cultivated varieties selected in East Africa in the early twentieth century: SL-09 and SL-34. SL-09 is of unknown origin while SL-34 descends from an heterogeneous “French Mission” population made of seeds brought by French Fathers of the Holy Ghost directly from Yemen (Aden). We named this cluster the “Yemen SL-34” cluster. The other cluster included Yemeni accessions and various worldwide cultivated varieties including Bourbon, Typica, SL28 (which has the same DNA fingerprinting as Coorg), Kent, KP-263 and 532, Bronze 009 and Moka. We named this cluster the “Yemen Typica-Bourbon” cluster. Typica and Bourbon are the only two varieties that spread into the world since the eighteenth century: Typica through India and then Asia and Bourbon via the Bourbon Island, now the La Reunion Island. Kent and Coorg are Indian selections deriving from Old Chicks, the population formed by the first introduction of coffee in India by Baba Budan (Haarer 1958; Kushalappa and Eskes 1989). In East Africa, the denomination “Moka” was given to any coffee beans or seeds proceeding from the port of Mocha in Yemen (Jones 1956). Cramer (1957) and Carvalho et al. (1984) described the Moka variety as having small roundish fruits, originating from a mutation than can occur in different genetic backgrounds.
Finally, we found one cluster which was made only of Yemeni samples and no other worldwide cultivars. We named this the “New-Yemen” cluster.
Figure 2 shows the graph based on the first two axis of the PCoA, which explained 40.2% of the whole variation. The “Ethiopian only” genetic cluster is on the right part of the graph while “SL-17”, “Yemen Bourbon/Typica” and “Yemen SL-34” are in the upper left quarter. Only New-Yemen is on the lower left part of the graph, thus confirming its genetic singularity.
The average allele number per marker was 7.4 for the whole population. However, the “Ethiopian only” cluster had 7.0 alleles per marker while all the other clusters together had only 2.63 alleles per marker. “SL-17”, “Yemen Bourbon/Typica”, “Yemen SL-34” and “New-Yemen” clusters had 3.5, 2.4, 1.8 and 1.9 alleles per marker, respectively.
The DA based on the coordinates on the five first axis confirmed the statistical difference between the clusters as the Wilks lambda test was highly significant (P < 0.0001). The overall good classification through cross-validation of samples to genetic clusters averaged 91%, and was 100% for the “New-Yemen” cluster.
The detailed list of samples with their sample category and attributed genetic cluster is provided in supplemental data (Table S1).
Discussion
To the best of our knowledge, our study is the first ever to zoom in on the genetic diversity of C. arabica in Yemen. Unveiling this genetic diversity enables a better understanding of the genetic diversity of the coffee varieties cultivated worldwide.
The relevance of the set of SSR markers used in the present study was confirmed for the genetic diversity exploration of C. arabica, as Pruvot-Woehl et al. (2020) established it. This was also in agreement of the usefulness of SSRs in genetic studies (Hodel et al. 2016). The set of 8 SRR markers is highly polymorphic with 7.4 alleles per marker in our study. Pruvot-Woehl et al. (2020) reported 11.9 alleles per marker for the same eight markers. However, introgressed varieties (descending from interspecific crosses with C. canephora or C. liberica) were included in the studied population. Da Silva et al. (2019) also included introgressed varieties and even other Coffea species in their study and had 6.9 alleles per marker (30 SSRs), hence less than in the present study. Benti et al. (2020) found 7.5 alleles per marker (14 SSRs) for a population of 40 Ethiopian Arabica varieties, two of which were introgressed varieties.
The DA proved the robustness of the genetic clusters, namely of the “New-Yemen” cluster.
Yemen holds most of the C. arabica genetic diversity known outside of Ethiopia
Our study confirmed previous knowledge based on history (de la Roque 1716; Ukers 1922; Chevalier 1929; Cramer 1957; Haarer 1958); Meyer 1965; Koehler 2017) and past genetic studies (Lashermes et al. 1996; Anthony et al. 2002; Silvestrini et al. 2007; Pruvot-Woehl et al. 2020; Scalabrin et al. 2020) that the vast majority of the coffee varieties cultivated outside Ethiopia transited through Yemen. However, the detailed genetic structure of the coffee trees cultivated in Yemen and the connection of this detailed genetic diversity to the varieties cultivated worldwide was not known. We have shown in this study that there are at least three distinct C. arabica genetic clusters in Yemen. Those clusters are well separated, leading to three main hypotheses: (i) Introduction of several populations from different narrow genetic basis and selection along time of the most adapted populations, (ii) introduction of one or several populations with a larger genetic basis and selection in Yemen within those populations of the different genetic clusters observed today or (iii) reintroduction in Yemen of the varieties selected worldwide.
Our data could support either of the first two hypothesis, or a combination of the two. The third hypothesis cannot be dismissed but is unlikely because there is no record in history of such re-introduction of all major coffee varieties back to Yemen.
The Yemen Typica-Bourbon cluster encompasses most of the important varieties cultivated worldwide. Hence, Yemen today is still holding most of the genetic diversity that it delivered to the world 300 years ago. Moreover, Yemen also hosts a unique specific genetic diversity. Indeed, no world wide cultivated varieties in our study belongs to the New-Yemen cluster, meaning that either it spread out of Yemen in the eighteenth century but was lost or counter-selected en route or it simply never left Yemen.
There was no correlation between the genetic clusters and the name of coffee cultivars given by Yemeni coffee farmers. This was in line with the findings of Hussein et al. (2017). In fact, most of the given names are related to some obvious visual characteristics that are not dependent on the precise genetic background. For instance, taller coffees would be called Udaini, Jufaini and Jadi while more compact trees would be called Dawairi and Tufahi (Sheibani, own observation). The discrepancy between given names and actual genetic fingerprint has been precisely shown by Pruvot-Woehl et al. (2020) in various parts of the world.
Origin and history of the C. arabica coffee varieties cultivated worldwide
None of the Ethiopian accessions in our study clustered with the Yemeni accessions. However, Ethiopian accessions samples are not representative of all the possible existing genetic diversity in Ethiopia (FAO 1968; Charrier 1978; Scalabrin et al. 2020). Furthermore, the only available Ethiopian germplasm—used in this study—comes from two surveys made some 50 years ago that did not cover the South Sudan region and the East Ethiopian Hararghe coffee zones. Scalabrin et al. (2020) suggest that there has been a general West–East movement of C. arabica from South West Ethiopia/South Sudan towards the Ethiopian East part of the Great Rift, then to Hararghe Ethiopian coffee zones, then to Yemen and then to the world. Montagnon and Bouharmont (1996) were the first to highlight a genetic difference between the Ethiopian accessions West and East of the Great Rift Valley. Whether the East of the Great Rift was home to wild C. arabica or whether it was a first place of domestication with wild C. arabica coming from the Western forests remains an open question. Further East, Hararghe zones were planted with coffee trees coming from the West and/or Eastern parts of the Great Rift Valley. Hararghe coffee could be related to coffee planted in Yemen as there were intense trade relationship between the two regions (Haarer 1958). Scalabrin et al. (2020) focusing on Ethiopian accessions identified three main genetic groups: the ‘Jimma-Bonga’ and ‘Sheka’ groups were made of Ethiopian accessions from the Western part of the coffee areas in Ethiopia while the third group, called “Harar-Yemen”, was more closely related to the worldwide cultivars and the Yemeni samples part of their study. Our study, through focusing on the Yemen coffee genetic diversity, indicated that Yemen accessions are made of genetically distinct clusters and that their likely origin in Ethiopia is not a single one. Under this scenario, studying more Ethiopian samples coming from the Eastern part of the Great Rift Valley and from Hararghe is a priority to better understand the origin of the Yemeni coffee genetic landscape.
The worldwide cultivars found in the Ethiopian Only cluster of our study represent coffee seeds that did not pass through the Yemen route. Geisha (CATIE code T.02722) is a referenced famous example. It was brought out of South Western Ethiopia directly to Kenya in the early twentieth century (Koehler 2017). The Java variety is another example. It was selected from an “Abyssinian”, hence Ethiopian, population, that traveled first to Indonesia and then to Cameroon where the Java variety was finally selected (Bouharmont 1994). SL-06 was also found in the Ethiopian Only Cluster, yet was first reported by Jones (1956) to be a single tree selection from Kent. Clearly clustering with Ethiopian landraces and genetically distinct from any Yemeni clusters, it is very unlikely that SL-06 was part of the seeds that transited to India and were at the origin of Kent. We cannot rule out potential mislabeling or mixing anywhere between the Kenyan research stations and the CATIE germplasm collection, so this “SL-06” might be unrelated to the original SL-06 Kenyan Selection. Chiroso is a variety cultivated at small scale in Colombia, famous for its cup quality. We show in our study that Chiroso is part of those Ethiopian landraces that “escaped” Ethiopia. It is very likely that the more DNA fingerprints collected of varieties grown in small scale with exceptional cup quality (Montagnon et al. 2019; Pruvot-Woehl et al. 2020), the more examples of Ethiopian landraces that bypassed the Yemen route will be found. Mibirizi CATIE code T.02702 is part of this “Ethiopian Only cluster” and is genetically very close to Ethiopian accession T.04620 of the CATIE collection. Another Mibirizi accession with CATIE code T.03622 is part of the SL-17 cluster and is genetically very different from Mibirizi with CATIE code T.02702. This might be due to mislabeling for the Mibirizi variety somewhere between East Africa and the CATIE collection. Given the limited information on Mibirizi (Leplae 1936), it is very unlikely that Mibirizi is an Ethiopian landrace and if any of the two Mibirizi codes is correct, it would rather be the T.03622 code. However, the hypothesis can’t be discarded that several independent introductions in Central Africa, through trade or cultural contact, led to two genetic backgrounds of the material both called Mibirizi.
In our study, the SL-17 cluster includes four Ethiopian accessions and no Yemeni accessions. Hence, one hypothesis is that it transited through Yemen but was not captured in the Yemen Qima population either because representatives of this cluster still exist but were not surveyed or because it has disappeared in Yemen. The other hypothesis is that it never transited through Yemen and was directly introduced in East Africa from Ethiopia.
The Yemen SL-34 cluster, unlike the SL-17 cluster, is clearly represented in Yemen. The two worldwide cultivated varieties part of this cluster are SL-09 and SL-34. According to Jones (1956), SL-09 is of unknown origin while SL-34 if from the “French Mission” origin, whose geographical origin is either Central Africa, Bourbon Island or Yemen.
The Yemen Typica-Bourbon represents the main early routes followed by the majority of C. arabica varieties cultivated worldwide. As reviewed by Montagnon et al. (2019) and Pruvot-Woehl et al. (2020), the two main coffee routes out of Yemen in the early eighteenth century were the Bourbon Island (today La Reunion) and India. The seeds that transited through the Bourbon Island most likely represented a small fraction of the Yemen Typica-Bourbon cluster as it only gave rise to the Bourbon variety. The seeds that transited through India necessarily included a wider diversity of the Yemen Typica-Bourbon as it gave rise to the Typica variety through the Indonesian route and then to the new world. These same population also gave rise to several varieties from this cluster that were first cultivated in India and then introduced to East Africa.
Finally, the New-Yemen cluster either never left Yemen or was lost en route. Regardless of which scenario is true, the fact is that it constitutes a unique genetic cluster not found in the cultivated varieties worldwide. The origins of this cluster in Ethiopia are unknown. Its ancestors in Ethiopia may have disappeared due to deforestation or genetic extinction. Similarly, it is also possible that its ancestors may exist in some populations in Ethiopia whose genetic diversity has not yet been studied or published.
Our study allows us to connect the dots and better understand the movement and spread of C. arabica around the world. Altogether, until today, three main routes were considered: (i) the Yemen-India route, (ii) the Yemen-Bourbon Island route and (iii) what we refer to as the “Escapee” route of some Ethiopian accessions that spread to the world without passing through Yemen. Our results confirm these three routes with the importance of the Yemen Typica-Bourbon cluster for the first two routes. However, it also highlights another previously overlooked route: the direct Yemen-East African route in the late 19th/early twentieth century, that the SL-17 and the Yemen SL-34 cluster might well have followed. The results also confirm the need for further exploration of the genetic diversity in the Hararghe region as a priority to investigate any connections between the Yemen clusters and their ancestors in Ethiopia.
Hence, for the first time, we present here deeper knowledge and a fuller picture of the genetic structure of C. arabica in Yemen, the gateway country between Ethiopia, the homeland of C. arabica coffee, and the world of cultivated coffee varieties. The varieties out of Yemen have been incredibly resilient: Bourbon and Typica have been cultivated for 300 years. Mundo Novo, Caturra or Catuai, all selected from the Yemen Typica-Bourbon varieties, have been successful in Brazil for more than 50 years now (Guerreiro Filho et al. 2018). SL-28, also from the Yemen Typica-Bourbon cluster is appreciated by Kenyan farmers for almost one century. Only the introgressed varieties originating from interspecific crosses with Robusta have partly taken over the original Yemen descending varieties for their yield and (now often compromised) resistance to coffee leaf rust (Zambolim 2016; Montagnon et al. 2019). Most recently, the F1 hybrids taking advantage of the hybrid vigor have shown a significant superiority to the traditional Yemen descending varieties (Georget et al. 2019; Marie et al. 2020).
Breseghello and Coelho (2013) proposed a comprehensive review of events from crop domestication to modern breeding of crops. Just after domestication, “the origin of crop”, comes the intuitive farmer selection, which is the “origin of landraces”, followed by pure line selection and mass selection, constituting the “origin of cultivars”. Later on, plant breeding based on controlled mapping and possibly marker assisted selection will take place. Our results indicate that Yemen was at the very least a major origin of landraces and cultivars for the world, forming a cluster distinct from Ethiopian accessions. This general observation was made in former studies (Montagnon and Bouharmont 1996; Lashermes et al. 1996; Anthony et 2002; Silvestrini et al. 2007; Pruvot-Woehl et al. 2020; Scalabrin et al. 2020). However, the present study goes deeper in the description of the genetic diversity of C. arabica in Yemen and its significance for the subsequent selection of cultivars worldwide. This pattern of genetic distance between wild and cultivated populations as the result of domestication and early selection has been observed in annual (Gepts 2004; Glaszmann et al. 2010) as well as in perennial crops, namely in tea—Camellia sinensis (L.) Kuntze (Meegahakumbura et al. 2018), peach—Prunus persica (L.) Batsch (Agaki et al. 2016) or grapevine–Vitis vinifera L. (Riaz et al. 2018).
Gepts (2004) recalls that crop domestication is a long-term selection experiment that has genetic consequences and often decreases the genetic diversity and the gene expression diversity (Flint-Garcia 2013; Liu et al. 2019; Turner-Hissong et al. 2020). This in turn offers a unique opportunity for breeders to understand, identify and target genes for adaptation (Ross-Ibarra et al. 2007; Glaszmann et al. 2010), including for polygenic adaptation as recently shown in cocoa- Theobroma cacao L. (Hämälä et al. 2020).
With our results, it is now possible to revisit and fully explore Yemen genetic diversity. Yemen’s coffee land has a rough climate: displaying both high and low temperatures in the extreme range of coffee growing areas worldwide, together with one of the lowest global rainfall levels. There is no doubt that this environment has favored resilient varieties, not only between the 1400s (coffee first introduced of Yemen) and 1700s (when today’s main worldwide coffee varieties were taken out of Yemen), but also during the last 300 years of coffee cultivation and propagation. We also unveil the New-Yemen cluster that has not been observed anywhere else in the world so far. This newly found genetic cluster represents a huge opportunity for the sustainability of the global coffee sector. Indeed, addressing the effects of climate change in coffee (Bunn et al. 2015; Davis et al. 2019) will partly rely on new varieties adapted to extreme temperatures. Yemen can offer the world of coffee several centuries worth of extreme coffee climate selected genes. The findings could not only provide the global coffee community with a deeper exploration and understanding of the genetic diversity at the origin of proven successful varieties but also offer a completely new genetic reservoir: the New-Yemen cluster.
In addition to the challenge of climate change, our results offer the specialty coffee market new unexplored genetic diversity for cup quality; which can significantly increase the diversity and sustainability of the coffee sector (Montagnon et al. 2019).
Last but not least, while Yemen is one of the oldest coffee growing countries, very little was known about the country’s coffee genetic landscape. Our results will be critical in guiding the selection of the best planting material for the Yemeni coffee growers.
Conclusion
To the best of our knowledge, our study is the first to unveil and describe the genetic diversity of C. arabica in Yemen, a key center for the development of the cultivated varieties around the world. We observed three genetic clusters in Yemen. Hence, either coffee was introduced in Yemen in a single event with a genetic diversity covering the Yemeni three clusters, or there were several independent introductions of coffee in Yemen. Furthermore, we showed that the major part of the genetic diversity of the coffee cultivars is still present today, 300 years after coffee was propagated out of Yemen to be cultivated around the world. However, one genetic cluster, the New-Yemen cluster, was not known before our study as it has not been observed elsewhere in the world, either because it never left Yemen or because it was lost en route. The New-Yemen cluster is not related to any population in Ethiopia observed thus far from a genetic point of view. Hence, either the genetic source was lost in Ethiopia or it is related to populations that have not been yet included in genetic diversity analysis. This work paves the way to new research opportunities, namely the search for adaptation genes in the Yemeni coffee gene pool.
Data availability
Data is available on demand.
References
Addinsoft (2020). XLSTAT statistical and data analysis solution. New York. USA. https://www.xlstat.com. Accessed 01 July 2020
Adhikari S, Saha S, Biswas A, Rana TS, Bandyopadhyay TK, Ghosh P (2017) Application of molecular markers in plant genome analysis: a review. Nucleus 60:283–297. https://doi.org/10.1007/s13237-017-0214-7
Akagi T, Hanada T, Yaegaki H, Gradziel TM, Tao R (2016) Genome-wide view of genetic diversity reveals paths of selection and cultivar differentiation in peach domestication. DNA Res 23:271–282. https://doi.org/10.1093/dnares/dsw014
Alkimim ER, Caixeta ET, Sousa TV, Resende MDV, da Silva FL, Sakiyama NS, Zambolim L (2020) Selective efficiency of genome-wide selection in Coffea canephora breeding. Tree Genet Gen. https://doi.org/10.1016/j.molp.2015.02.002
Al-Murish TM, Elshafei AA, Al-Doss AA, Barakat MN (2013) Genetic diversity of coffee (Coffea arabica L.) in Yemen via SRAP, TRAP and SSR markers. J Food Agric Environ 11:411–416
Anthony F, Combes MC, Astorga C, Bertrand B, Graziosi G, Lashermes P (2002) The origin of cultivated Coffea arabica L. varieties revealed by AFLP and SSR markers. Theor Appl Genet 104:894–900. https://doi.org/10.1007/s00122-001-0798-8
Avelino J, Cristancho M, Georgiou S et al (2015) The coffee rust crises in Colombia and Central America (2008–2013): impacts, plausible causes and proposed solutions. Food Secur 7:303–321. https://doi.org/10.1007/s12571-015-0446-9
Benti T, Gebre E, Tesfaye K, Berecha G, Lashermes P, Kyallo M, Kouadio Yao N (2020) Genetic diversity among commercial arabica coffee (Coffea arabica L.) varieties in Ethiopia using simple sequence repeat markers. J Crop Improv. https://doi.org/10.1080/15427528.2020.1803169
Bouharmont P (1994) La variété Java: un caféier arabica sélectionné au Cameroun. Plantations, recherche, développement 1:38–45
Breseghello F, Coelho ASG (2013) Traditional and modern plant breeding methods with examples in rice (Oryza sativa L.). J Agric Food Chem 61:8277–8286. https://doi.org/10.1021/jf305531j
Browning D (2018). How many coffee farms are there in the world?. ASIC Conference Portland, 2018/09/16–20 https://www.youtube.com/watch?v=vKaeDkpqPSg, Accessed 01 July 2020
Bunn C, Läderach P, Jimenez JGP, Montagnon C, Schilling T (2015) Multiclass classification of agro-ecological zones for Arabica coffee: an improved understanding of the impacts of climate change. PLoS ONE. https://doi.org/10.1371/journal.pone.0140490
Carvalho A, Fazuoli LC, Medina Filho HP (1984) Effects of X-radiation on the induction of mutations in Coffea arabica. Bragantia 43:553–567
Charrier A (1978). Etude de la structure et de la variabilité génétique des caféiers : Résultats des études et des expérimentations réalisées au Cameroun, en Côte d’Ivoire et à Madagascar sur l’espèce Coffea arabica L. collectée en Ethiopie par une mission Orstom en 1966. Bulletin IFCC n° 14, Paris, FRA.
Chevalier A (1929) Les caféiers du globe. I. Généralités sur les caféiers, Encyclopédie biologique, Paul Lechevalier, Paris
Clevenger J, Chavarro C, Pearl SA, Ozias-Akins P, Jackson SA (2015) Single nucleotide polymorphism identification in polyploids: a review, example, and recommendations. Mol plant 8:831–846. https://doi.org/10.1016/j.molp.2015.02.002
Combes MC, Andrzejewski S, Anthony F, Bertrand B, Rovelli P, Graziosi G, Lashermes P (2000) Characterization of microsatellite loci in Coffea arabica and related coffee species. Mol Ecol 9:1178–1180. https://doi.org/10.1046/j.1365-294x.2000.00954-5.x
Cramer P J S (1957). A Review of Literature of Coffee Research in Indonesia (from about 1602 to 1945). IICA.
da Silva BSR, Sant’Ana G C, al, (2019) Population structure and genetic relationships between Ethiopian and Brazilian Coffea arabica genotypes revealed by SSR markers. Genetica 147:205–216. https://doi.org/10.1007/s10709-019-00064-4
Davis AP, Chadburn H, Moat J, O’Sullivan R, Hargreaves S, Lughadha EN (2019) High extinction risk for wild coffee species and implications for coffee sector sustainability. Sci Adv. https://doi.org/10.1126/sciadv.aav3473Davis
De La Roque J (1716) Voyage de l'Arabie heureuse, par l'Océan oriental, et le détroit de la Mer Rouge : fait par les François pour la première fois, dans les années 1708, 1709 et 1710. André Cailleau, Paris. https://play.google.com/books/reader?id=3fsOAAAAQAAJ&hl=fr&num=10&printsec=frontcover&pg=GBS.PP7. Accessed 01 July 2020.
Emanuelli F, Lorenzi S, Grzeskowiak L et al. (2013). Genetic diversity and population structure assessed by SSR and SNP markers in a large germplasm collection of grape. BMC plant biology http://www.biomedcentral.co m/1471–2229/13/39
FAO (1968) FAO coffee mission to Ethiopia: 1964–1965. FAO, Rome
Fernie LM (1970) The improvement of arabica coffee in East Africa. Crop Improvement in East Africa. Techn, Comm, p 19
Flint-Garcia SA (2013) Genetics and consequences of crop domestication. J Agric Food Chem 61:8267–8276. https://doi.org/10.1021/jf305511d
Georget F, Marie L, Alpizar E et al (2019) Starmaya: The first arabica F1 coffee hybrid produced using genetic male sterility. Front Plant Sci 10:1344. https://doi.org/10.3389/fpls.2019.01344/full
Gepts P (2004) Crop domestication as a long-term selection experiment. Plant Breed Rev 24:1–44
Glaszmann JC, Kilian B, Upadhyaya HD, Varshney RK (2010) Accessing genetic diversity for crop improvement. Curr Opin Plant Biol 13:167–173. https://doi.org/10.1016/j.pbi.2010.01.004
Guerreiro Filho O, Ramalho MAP, Andrade VT (2018) Alcides Carvalho and the selection of Catuaí cultivar: interpreting the past and drawing lessons for the future. Crop Breed and Appl Biotechnol 18:460–466
Haarer AE (1958) Modern coffee production. Ebenezer Baylis and Son, The Trinity Press, London (UK)
Hämälä T, Guiltinan MJ, Marden JH, Maximova SN, dePamphilis CW, Tiffin P (2020) Gene expression modularity reveals footprints of polygenic adaptation in Theobroma cacao. Mol Biol Evol 37:110–123. https://doi.org/10.1093/molbev/msz206
Harlan JR (1969) Ethiopia: a center of diversity. Econ Bot 23:309–314
Hodel RG, Segovia-Salcedo MC, Landi JB et al (2016) The report of my death was an exaggeration: a review for researchers using microsatellites in the 21st century. Appl Plant Sci. https://doi.org/10.3732/apps.1600025
Hussein MAA, Al-Azab AAA, Habib SS, El Sherif FM, El-Garhy HA (2017) Genetic diversity, structure and DNA fingerprint for developing molecular IDs of Yemeni coffee (Coffea Arabica L.) Germplasm assessed by SSR Markers. Egypt J Plant Breed 203:1–25
Jones P A (1956). Notes on the varieties of Coffea arabica in Kenya. Coffee Board of Kenya Monthly Bulletin, November 1956.
Koehler J (2017) Where the wild coffee grows: The untold story of coffee from the cloud forests of Ethiopia to your cup. Bloomsbury Publishing, USA
Kushalappa AC, Eskes AB (1989) Advances in coffee rust research. Annu Rev Phytopathol 27:503–531. https://doi.org/10.1146/annurev.py.27.090189.002443
Lashermes P, Trouslot P, Anthony F, Combes M, Charrier A (1996) Genetic diversity for RAPD markers between cultivated and wild accessions of Coffea arabica. Euphytica 87:59–64
Leplae E (1936) Les plantations de café au Congo belge, leur histoire, 1881–1935, leur importance actuelle. Van Campenhout G, Bruxelles
Liu W, Chen L, Zhang S et al (2019) Decrease of gene expression diversity during domestication of animals and plants. BMC Evol Biol. https://doi.org/10.1186/s12862-018-1340-9
Marie L, Abdallah C, Campa C et al (2020) G× E interactions on yield and quality in Coffea arabica: new F1 hybrids outperform American cultivars. Euphytica 216:1–17. https://doi.org/10.1007/s10681-020-02608-8
Meegahakumbura MK, Wambulwa MC, Li MM et al (2018) Domestication origin and breeding history of the tea plant (Camellia sinensis) in China and India based on nuclear microsatellites and cpDNA sequence data. Front Plant Sci. https://doi.org/10.3389/fpls.2017.02270
Meyer FG (1965) Notes on wild Coffea arabica from Southwestern Ethiopia, with some historical considerations. Econ Bot 19:136–151
Montagnon C, Bouharmont P (1996) Multivariate analysis of phenotypic diversity of Coffea arabica. Genet Resour Crop Evol 43:221–227
Montagnon C, Marraccini P, Bertrand B (2019) Breeding for coffee quality. In: Oberthur et al. (eds) Specialty Coffee-Managing Quality. Cropster Innsbruck, Austria, pp 109–143
Perrier X, Jacquemoud-Collet J P (2006). DARwin software. http://darwin.cirad.fr/darwin
Pruvot-Woehl S, Krishnan S, Solano W, Schilling T, Toniutti L, Bertrand B, Montagnon C (2020) Authentication of Coffea arabica varieties through DNA fingerprinting and its significance for the coffee sector. J AOAC Int 103:325–334. https://doi.org/10.1093/jaocint/qsz003
Riaz S, De Lorenzis G, Velasco D et al (2018) Genetic diversity analysis of cultivated and wild grapevine (Vitis vinifera L.) accessions around the Mediterranean basin and Central Asia. BMC Plant Biol. https://doi.org/10.1186/s12870-018-1351-0
Ross-Ibarra J, Morrell PL, Gaut BS (2007) Plant domestication, a unique opportunity to identify the genetic basis of adaptation. Proc Natl Acad Sci 104:8641–8648. https://doi.org/10.1073/pnas.0700643104
Saitou N, Nei M (1987) The Neighbor-Joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Scalabrin S, Toniutti L, Di Gaspero G et al. (2020). A single polyploidization event at the origin of the tetraploid genome of Coffea arabica is responsible for the extremely low genetic variation in wild and cultivated germplasm. Scientific Reports.
Silvestrini M, Junqueira MG, Favarin AC, Guerreiro-Filho O, Maluf MP, Silvarolla MB, Colombo CA (2007) Genetic diversity and structure of Ethiopian, Yemen and Brazilian Coffea arabica L. accessions using microsatellites markers. Genet Resour Crop Evol 54:1367–1379. https://doi.org/10.1007/s10722-006-9122-4
Singh N, Choudhury DR, Singh AK et al (2013) Comparison of SSR and SNP markers in estimation of genetic diversity and population structure of Indian rice varieties. PLoS ONE. https://doi.org/10.1371/journal.pone.0084136
Sylvain PG (1958) Ethiopian coffee—its significance to world coffee problems. Econ Bot 12:111–139
Thomas AS (1942) The wild Arabica coffee on the Boma Plateau. Anglo-Egyptian Sudan Emp J Exp Agric 10:207–212
Tomassone R, Danzart M, Daudin J J and Masson J P (1988). Discrimination et classement. Masson.
Turner-Hissong SD, Mabry ME, Beissinger TM, Ross-Ibarra J, Pires JC (2020) Evolutionary insights into plant breeding. Curr Opin Plant Biol 54:93–100. https://doi.org/10.1016/j.pbi.2020.03.003
Ukers MA (1922) All about coffee. The Tea and Coffee Trade Journal, New York
Van der Vossen H, Bertrand B, Charrier A (2015) Next generation variety development for sustainable production of arabica coffee (Coffea arabica L.): a review. Euphytica 204:243–256. https://doi.org/10.1007/s10681-015-1398-z
Zambolim L (2016) Current status and management of coffee leaf rust in Brazil. Trop Plant Pathol 41:1–8. https://doi.org/10.1007/s40858-016-0065-9
Acknowledgements
Authors would like to thank Dr Jane Cheserek from the Kenya Agricultural & Livestock Research Organization who found and provided us with the original article of Jones (1956) “Notes on the varieties of Coffea arabica in Kenya”, as well as two anonymous reviewers for their useful comments. Authors would also like to thank the Ministry of Agriculture and Irrigation of Yemen for their support of the work and the coffee farmers of Yemen.
Funding
Qima Coffee.
Author information
Authors and Affiliations
Contributions
CM wrote the paper and did all the genetic analysis, MA oversaw the breeding populations in Yemen, WS provided data for Ethiopian accessions and participated in the interpretation/discussion of data, FS had the idea of the study, prepared the samples from Yemen and participated to the discussion and writing of the paper.
Corresponding author
Ethics declarations
Conflict of interest
The authors declares that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Montagnon, C., Mahyoub, A., Solano, W. et al. Unveiling a unique genetic diversity of cultivated Coffea arabica L. in its main domestication center: Yemen. Genet Resour Crop Evol 68, 2411–2422 (2021). https://doi.org/10.1007/s10722-021-01139-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-021-01139-y