Abstract
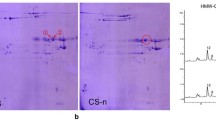
High molecular weight glutenin subunits (HMW-GSs) are the critical components of wheat seed storage proteins. They play a crucial role in determining dough viscoelastic properties and end-use quality of wheat flour. To investigate the potential utilization of the special HMW-GSs from wild emmer wheat (2n = 4x = 28, AABB) accession D141 in common wheat (2n = 6x = 42, AABBDD), continued to our previous report of the 1Ay and its coding sequence, the other three HMW-GSs, 1Ax, 1Bx13.1 and 1By16.1, and their coding genes were isolated, characterized and transferred from D141. The 1Ax gene was silent because of containing a premature stop codon TGA at position 835–837. The ORFs of 1Bx13.1 and 1By16.1 were 2373 and 2157 bp in length, and encoded 789 and 717 amino acid residues, respectively. Compared with the other 20 Glu-1Bx alleles in GenBank, seven SNPs and one 18-bp deletion were detected in 1Bx13.1. Compared with the other 10 Glu-1By alleles in GenBank, nine SNPs were observed in 1By16.1. The mobility of 1Bx13.1 and 1By16.1 on the SDS-PAGE gel was most similar to that of 1Bx13 and 1By16 from common wheat (T. aestivum, 2n = 6x = 42, AABBDD) cv. ‘Jimai 20′, respectively. Protein secondary structure prediction showed that the combination of 1Bx13.1 + 1By16.1 possessed more α-helixes with more amino acid residues than that of 1Bx13 + 1By16. It suggested that 1Bx13.1 + 1By16.1 from wild emmer accession D141 might have more positive effects on bread-making quality than the superior subunit combination 1Bx13 + 1By16. The HMW-GS composition analysis of F1 and F2 between common wheat for female and wild emmer accession D141 for male, indicated that the special HMW-GSs from wild emmer accession D141 could be easily introduced into common wheat (2n = 42) by general inter-species hybridization.






Similar content being viewed by others
Abbreviations
- aa:
-
Amino acid(s)
- bp:
-
Base pair(s)
- CTAB:
-
Cetyltrimethyl ammonium bromide
- dNTP:
-
Deoxyribonucleoside triphosphate
- HMW-GS:
-
High molecular weight glutenin subunit
- InDel:
-
Insertion and deletion
- IPTG:
-
Isopropyl β-Δ-thiogalactopyranoside
- ORF:
-
Open reading frame
- PCR:
-
Polymerase chain reaction
- SDS-PAGE:
-
Sodium dodecyl sulfate polyacrylamide gel electrophoresis
- SNP:
-
Single nucleotide polymorphism
References
Anderson OD, Greene FC (1989) The characterization and comparative analysis of high-molecular-weight glutenin genes from genomes A and B of a hexaploid wheat. Theor Appl Genet 77:689–700
Anjum FM, Khan MR, Din A, Saeed M, Pasha I, Arshad MU (2007) Wheat gluten: high molecular weight glutenin subunits—structure, genetics, and relation to dough elasticity. J Food Sci 72:R56–R63
Bi ZG, Wu BH, Hu XG, Guo XH, Liu DC, Zheng YL (2014) Identification of an active 1Ay gene from Triticum turgidum ssp. dicoccoides. Czech J Genet Plant Breed 50:208–215
Ciaffi M, Lafiandra D, Porceddu E, Benedettellim S (1993) Storage protein variation in wild emmer wheat (Triticum turgidum ssp. dicoccoides) from Jordan and Turkey. I. Electrophoretic characterization of genotypes. Theor Appl Genet 86:474–480
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Forde J, Malpica JM, Halford NG, Shewry PR, Anderson OD, Greene FC, Miflin BJ (1985) The nucleotide sequence of a HMW glutenin subunit gene located on chromosome 1A of wheat (Triticum aestivum L.). Nucleic Acids Res 13:6817–6832
Gianibelli MC, Larroque OR, MacRichie F, Wrigley CW (2001) Biochemical, genetic, and molecular characterization of wheat glutenin and its component subunits. Cereal Chem 78:635–646
Guo XH, Bi ZG, Wu BH, Wang ZZ, Hu JL, Zheng YL, Liu DC (2013a) ChAy/Bx, a novel chimeric high-molecular-weight glutenin subunit gene apparently created by homoeologous recombination in Triticum turgidum ssp. dicoccoides. Gene 531:318–325
Guo XH, Wu BH, Hu XG, Bi ZG, Wang ZZ, Liu DC, Zheng YL (2013b) Molecular characterization of two y-type high molecular weight glutenin subunit alleles 1Ay12 * and 1Ay8 * from cultivated einkorn wheat (Triticum monococcum ssp. monococcum). Gene 516:1–7
Halford NG, Field JM, Blair H, Urwin P, Moore K, Robert L, Thompson R, Flavell RB, Tatham AS, Shewry PR (1992) Analysis of HMW glutenin subunits encoded by chromosome 1A of bread wheat (Triticum aestivum L.) indicates quantitative effects on grain quality. Theor Appl Genet 83:373–378
Hu XG, Wu BH, Yan ZH, Liu DC, Wei YM, Zheng YL (2010) Characterization of a Novel 1Ay Gene and Its Expression Protein in Triticum urartu. Agr Sci China 9:1543–1552
Hu XG, Wu BH, Bi ZG, Liu DC, Zhang LQ, Yan ZH, Wei YM, Zheng YL (2012) Allelic variation and distribution of HMW glutenin subunit 1Ay in Triticum species. Genet Resour Crop Ev 59:491–497
Jiang QT, Wei YM, Wang F, Wang JR, Yan ZH, Zheng YL (2009) Characterization and comparative analysis of HMW glutenin 1Ay alleles with differential expressions. BMC Plant Biol 9:16
Jin M, Xie ZZ, Ge P, Li J, Jiang SS, Subburaj S, Li XH, Zeller FJ, Hsam SL, Yan YM (2012) Identification and molecular characterisation of HMW glutenin subunit 1By16 * in wild emmer. J Appl Genet 53:249–258
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Levy AA, Galili G, Feldman M (1988) Polymorphism and genetic control of high molecular weight glutenin subunits in wild tetraploid wheat Triticum turgidum var. dicoccoides. Heredity 61:63–72
Margiotta B, Urbano M, Colaprico G, Turchetta T, Lafiandra D (1998) Variation of high molecular weight glutenin subunits in tetraploid wheats of genomic formula AAGG. In: Slinkard AE (ed) Proceedings of the 9th international wheat genetics symposium. University Extension Press, Saskatoon, pp 195–197
McGuffin LJ, Bryson K, Jones DT (2000) The PSIPRED protein structure prediction server. Bioinformatics 16:404–405
Nevo E, Payne PI (1987) Wheat storage proteins: diversity of HMW glutenin subunits in wild emmer from Israel. 1. Geographical patterns and ecological predictability. Theor Appl Genet 74:827–836
Nevo E, Korol AB, Beiles A, Fahima T (2002) Evolution of wild emmer and wheat improvement: population genetics, genetic resources, and genome organization of wheat’s progenitor. Springer, Triticum dicoccoides
Pang BS, Zhang XY (2008) Isolation and molecular characterization of high molecular weight glutenin subunit genes 1Bx13 and 1By16 from hexaploid wheat. J Integr Plant Biol 50:329–337
Payne PI (1987) Genetics of wheat storage proteins and the effect of allelic variation on bread-making quality. Annu Rev Plant Physiol 38:141–153
Payne PI, Holt LM, Law CN (1981) Structural and genetical studies on the highmolecular-weight subunits of wheat glutenin. Part 1: allelic variation in subunits amongst varieties of wheat (Triticum aestivum). Theor Appl Genet 60:229–236
Payne PI, Lawrence GJ (1983) Catalogue of alleles for the complex gene loci. Glu-A1, Glu-B1 and Glu-D1 which code for high molecular weight subunits of glutenin in hexaploid wheat. Cereal Res Commun 11:29–35
Ribeiro M, Bancel E, Faye A, Dardevet M, Ravel C, Branlard G, Igrejas G (2013) Proteogenomic characterization of novel x-type high molecular weight glutenin subunit 1Ax1.1. Int J Mol Sci 14:5650–5667
Shewry PR, Tatham AS (1997) Disulphide bonds in wheat gluten proteins. J Cereal Sci 25:207–227
Shewry PR, Halford NG, Tatham AS (1992) High molecular weight subunits of wheat glutenin. J Cereal Sci 15:105–120
Shewry PR, Napier JA, Tatham AS (1995) Seed storage proteins: structures and biosynthesis. Plant Cell 7:945–956
Shewry PR, Halford NG, Belton PS, Tatham AS (2002) The structure and properties of gluten: an elastic protein from wheat grain. Philos Trans R Soc Lond B Biol Sci 357:133–142
Shewry PR, Gilbert SM, Savage AWJ, Tatham AS, Wan YF, Belton PS, Wellner N, D’Ovidio R, Békés F, Halford NG (2003) Sequence and properties of HMW subunit 1Bx20 from pasta wheat (Triticum durum) which is associated with poor end use properties. Theor Appl Genet 106:744–750
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Thompson RD, Bartels D, Harberd NP (1985) Nucleotide sequence of a gene from chromosome 1D of wheat encoding a HMW-glutenin subunit. Nucleic Acids Res 13:6833–6846
Wan Y, Wang D, Shewry PR, Halford NG (2002) Isolation and characterization of five novel high molecular weight subunit of glutenin genes from Triticum timopheevi and Aegilops cylindrica. Theor Appl Genet 104:828–839
Xie W, Nevo E (2008) Wild emmer: genetic resources, gene mapping and potential for wheat improvement. Euphytica 164:603–614
Xie RL, Wan YF, Zhang Y, Wang DW (2001) HMW glutenin subunits in multiploid Aegilops species: composition analysis and molecular cloning of coding sequences. Chin Sci Bull 46:309–313
Xu LL, Li W, Wei YM, Zheng YL (2009) Genetic diversity of HMW glutenin subunits in diploid, tetraploid and hexaploid Triticum species. Genet Resour Crop Evol 56:377–391
Xu SS, Khan K, Klindworth DL, Faris JD, Nygard G (2004) Chromosomal location of genes for novel glutenin subunits and gliadins in wild emmer wheat (Triticum turgidum L. var. dicoccoides). Theor Appl Genet 108:1221–1228
Yu WL, Li GJ, Wu BH, Zheng YL, Liu DC, Yan ZH (2005) Variation of high-molecular-weight glutenin subunits in Triticum dicoccoides and its potential utilization in common wheat. J Sichuan Agr Univ 23: 252-257 (in Chinese, with English abstract)
Conflict of interest
The authors declare there are no conflicts of interest regarding the publication of this paper.
Author information
Authors and Affiliations
Corresponding author
Additional information
Xiao-Hui Guo and Ji-Liang Hu have contributed equally to this work.
Rights and permissions
About this article
Cite this article
Guo, XH., Hu, JL., Wu, BH. et al. Special HMW-GSs and their genes of Triticum turgidum subsp. dicoccoides accession D141 and the potential utilization in common wheat. Genet Resour Crop Evol 63, 833–844 (2016). https://doi.org/10.1007/s10722-015-0287-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-015-0287-6