Abstract
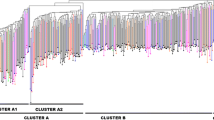
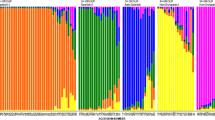
As a counterweight to genetic erosion, landraces could become important germplasm resources for breeding. In the present study the genetic diversity of 62 Greek and Eastern European oat accessions (seven cultivars and 55 common and red oat landraces collected from 1904 to 1960) was studied using simple sequence repeats and 31 morphological descriptors. High levels of polymorphism were detected and 209 distinctive alleles were identified; on average 14.65 alleles per primer. Both, the principal component analysis derived from the morphological data, and the unweighted pair group method with arithmetic mean from the molecular analysis clustered the oat genotypes according to their type (common and red oats). In addition, the analysis of molecular variation showed that there were noteworthy differences within designated geographic regions. Moreover, a more discrete subpopulation structure was identified based on allele frequency using a Bayesian clustering approach. A clear distinction was evident among the red and common oats suggesting that they should be considered as separate species.



Similar content being viewed by others
References
Banga O (1944) Veredeling van tuinbouwgewassen. Zwolle, pp 211
Baohong G, Zhou X, Murphy JP (2003) Genetic variation within Chinese and western cultivated oat accessions. Cereal Res Commun 31:339–346
Baum BR (1977) Oats: wild and cultivated, a monograph of the genus Avena L. (Poaceae). Canada Department of Agriculture, Ottawa
Boczkowska M, Tarczyk E (2013) Genetic diversity among Polish landraces of common oat (Avena sativa L.). Genet Resour Crop Evol 60:2157–2169
Boczkowska M, Nowosielski J, Nowosielska D, Podyma W (2014) Assessing genetic diversity in 23 early Polish oat cultivars based on molecular and morphological studies. Genet Resour Crop Evol 61:927–941
Coffman FA (1961) Origin and history. In: Coffman FA (ed) Oats and oat improvement. American Society of Agronomy, Madison, pp 15–40
Coffman FA (1977) Oat history, identification and classification. USDA-ARS Tech. Bull. No. 1516. U.S. Gov. Print. Office, Washington, DC
Danert S (1972) Zur Systematik von Avena sativa L. und zur Beurteilung von Avena nuda Hoj. [Taxonomy of Avena sativa L. and interpretation of Avena nuda Hojer]. Kulturpflanze 19:45–51 (in German)
Diederichsen A (2004) Case studies for the use of infraspecific classifications in managing germplasm collections of cultivated plants. Acta Hortic 634:127–139
Diederichsen A (2008) Assessments of genetic diversity within a world collection of cultivated hexaploid oat (Avena sativa L.) based on qualitative morphological characters. Genet Resour Crop Evol 55:419–440
Diederichsen A (2009) Duplication assessments in Nordic Avena sativa accessions at the Canadian national genebank. Genet Resour Crop Evol 56:587–597
Fu YB, Peterson GW, Scoles G, Rossnagel B, Schoen DJ, Richards KW (2003) Allelic diversity changes in 96 Canadian oat varieties released from 1886 to 2001. Crop Sci 43:1989–1995
Fu YB, Kibite S, Richards KW (2004) Amplified fragment length polymorphism analysis of 96 Canadian oat varieties released between 1886 and 2001. Can J Plant Sci 84:23–30
Fu YB, Peterson GW, Williams D, Richards KW, Fetch MJ (2005) Patterns of AFLP variation in a core subset of cultivated hexaploid oat germplasm. Theor Appl Genet 111:530–539
Hammer K, Gladis T, Diederichsen A (2003) In situ and on-farm management of plant genetic resources. Eur J Agron 19:509–517
He X, Bjørnstad A (2012) Diversity of North European oat analyzed by SSR, AFLP and DArT markers. Theor Appl Genet 125:57–70
IBPGR (1985) Oat doscriptors. International Board for Plant Genetic Resources, Rome
Li CD, Rossnagel BG, Scoles GJ (2000) The development of oat microsatellite markers and their use in identifying relationships among Avena species and oat cultivars. Theor Appl Genet 101:1259–1268
Li RZ, Wang SW, Duan LS, Li ZH, Christoffers MJ, Mengistu LW (2007) Genetic diversity of wild oat (Avena fatua) populations from China and the United States. Weed Sci 55(2):95–101
Loskutov IG (1998) The collection of wild oat species of C.I.S. as a source of diversity in agricultural traits. Genet Resour Crop Evol 45:291–295
Loskutov IG (2008) On evolutionary pathways of Avena species. Genet Resour Crop Evol 55:211–220
Loskutov IG, Rines HW (2011) Avena. In: Kole C (ed) Wild crop relatives: genomic and breeding resources. Springer, Berlin, pp 109–183
Montilla-Bascón G, Sánchez-Martín J, Rispail N, Rubiales D, Mur L, Langdon T, Griffiths I, Howarth C, Prats E (2013) Genetic diversity and population structure among oat cultivars and landraces. Plant Mol Biol Rep 31:1305–1314
Nersting L, Andersen S, von Bothmer R, Gullord M, Jørgensen R (2006) Morphological and molecular diversity of Nordic oat through one hundred years of breeding. Euphytica 150:327–337
Newell MA, Cook D, Tinker NA, Jannink JL (2011) Population structure and linkage disequilibrium in oat (Avena sativa L.): implications for genome-wide association studies. Theor Appl Genet 122:623–632
Newton AC, Akar T, Baresel JP, Bebeli PJ, Bettencourt E, Bladenopoulos KV, Czembor JH et al (2010) Cereal landraces for sustainable agriculture. A review. Agron Sustain Dev 30(2):237–269
Odonoughue LS, Souza E, Tanksley SD, Sorrells ME (1994) Relationships among North-American oat cultivars based on restriction-fragment-length-polymorphisms. Crop Sci 34:1251–1258
Pavlicek A, Hrda S, Flegr J (1999) FreeTree-freeware program for construction of phylogenetic trees on the basis of distance data and bootstrapping/jack-knife analysis of the tree robustness. Application in the RAPD analysis of the genus Frenkelia. Folia Biol (Praha) 45:97–99
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure from multilocus genotype data. Genetics 155:945–959
Rodionova NA, Soldatov VN, Merezhko VE, Jarosh NP, Kobyljanskij VL (1994) Flora of cultivated plants, vol 2, part 3, Oat. Kolos, Moscow
Sheikhehpour S, Bahraminejad S, Cheghamirza K (2014) Morphological and molecular genetic variations of oat genotypes grown in Kermanshah, Iran. Mol Biol Rep 41:4023–4030
Souza E, Sorrells ME (1991) Relationships among 70 North American oat germplasm: I. Cluster analysis using quantitative characters. Crop Sci 31:599–605
Tessier R, David J, This P, Boursiquot JM, Charrier A (1999) Optimization of the choice of molecular markers for varietal identification in Vitis vinifera L. Theor Appl Genet 98:171–177
Wesenberg DM, Briggle LW, Smith DH (1992) Germplasm collection, preservation and utilization. In: Marshall HG, Sorrells ME (eds) Oat science and technology. American Society of Agronomy, Madison, pp 793–820
Zohary D, Hopf M (2000) Domestication of plants in the Old World, 3rd edn. Oxford University Press, Oxford
Acknowledgments
This work was partially supported by the start-up funding of AK from CUT.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Nikoloudakis, N., Bladenopoulos, K. & Katsiotis, A. Structural patterns and genetic diversity among oat (Avena) landraces assessed by microsatellite markers and morphological analysis. Genet Resour Crop Evol 63, 801–811 (2016). https://doi.org/10.1007/s10722-015-0284-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-015-0284-9