Abstract
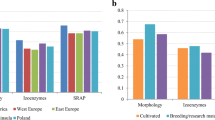
Oat is an important crop in Nordic countries both for feed and human consumption. Maintaining a high level of genetic diversity is essential for both breeding and agronomy. A panel of 94 oat accessions was used in this study, including 24 museum accessions over 100- to 120-year old and 70 genebank accessions from mainly Nordic countries and Germany, covering different breeding periods. Sixty-one polymorphic SSR, 201 AFLP and 1056 DArT markers were used to evaluate the past and present genetic diversity of the Nordic gene pool. Norwegian accessions showed the highest diversity, followed by Swedish and Finnish, with German accessions the least diverse. In addition, the Nordic accessions appeared to be highly interrelated and distinct from the German, reflecting a frequent germplasm exchange and interbreeding among Nordic countries. A significant loss of diversity happened at the transition from landraces and old cultivars to modern cultivars. Modern oat originated from only a segment of the landraces and left the remainder, especially black oat, unused. However, no significant overall diversity reduction was found during modern breeding periods, although fluctuation of diversity indices was observed. The narrow genetic basis of the modern Nordic gene pool calls for increasing genetic diversity through cultivar introduction and prebreeding based on neglected sources like the Nordic black oat.





Similar content being viewed by others
References
Achleitner A, Tinker NA, Zechner E, Buerstmayr H (2008) Genetic diversity among oat varieties of worldwide origin and associations of AFLP markers with quantitative traits. Theor Appl Genet 117:1041–1053
Åkerman Å, Granhall I, Nilsson-Leissner G, Müntzing A, Tedin O (1938) Swedish contributions to the development of plant breeding. A. Bonniers boktryckeri, Stockholm
Allaby RG, Jones MK, Brown TA (1994) DNA in charred wheat grains from the Iron Age hillfort at Danebury. Engl Antiq 68:126–132
Atterberg A (1887a) Försök till klassifikation af hafrevarieteterna efter deras kornformer. Kongl Lantbruksakademiens handlingar och tidskrift 26:135–160
Atterberg A (1887b) Kort Redogörelse öfver år 1886 insamlade och undersökta hafreprof. Kalmars Kemiska Anstalt, Kalmar
Becher R (2007) EST-derived microsatellites as a rich source of molecular markers for oats. Plant Breed 126:274–278
Bickelmann U (1989) The Pedigree of German oat cultivars (Avena sativa L.) and the occurrence of fatuoids. Plant Breed 103:163–170
Bjornstad A, Abay F (2010) Multivariate patterns of diversity in Ethiopian barleys. Crop Sci 50:1579–1586
Bjornstad A, Westad F, Martens H (2004) Analysis of genetic marker-phenotype relationships by jack-knifed partial least squares regression (PLSR). Hereditas 141:149–165
Brautigam M, Lindlof A, Zakhrabekova S, Gharti-Chhetri G, Olsson B, Olsson O (2005) Generation and analysis of 9792 EST sequences from cold acclimated oat, Avena sativa. BMC Plant Biol 5
Buerstmayr H, Krenn N, Stephan U, Grausgruber H, Zechner E (2007) Agronomic performance and quality of oat (Avena sativa L.) genotypes of worldwide origin produced under Central European growing conditions. Field Crops Res 101:343–351
Coffman FA (1977) Oat history, identification, and classification. Agricultural research service. US Department of Agriculture, Washington
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes—application to human mitochondrial-dna restriction data. Genetics 131:479–491
Frese L, Reinhard U, Bannier H-J, Germeier CU (2009) Landrace inventory in Germany—preparing the national implementation of the EU Directive 2008/62/EC. In: Veteläinen M, Negri V, Maxted N (eds) European landraces: on-farm conservation, management and use. Bioversity International, Rome, pp 79–96
Frey K (1998) Genetic responses of oat genotypes to environmental factors. Field Crops Res 56:183–185
Fu YB, Peterson GW, Scoles G, Rossnagel B, Schoen DJ, Richards KW (2003) Allelic diversity changes in 96 Canadian oat cultivars released from 1886 to 2001. Crop Sci 43:1989–1995
Fu YB, Kibite S, Richards KW (2004) Amplified fragment length polymorphism analysis of 96 Canadian oat cultivars released between 1886 and 2001. Can J Plant Sci 84:23–30
Fu YB, Chong J, Fetch T, Wang ML (2007) Microsatellite variation in Avena sterilis oat germplasm. Theor Appl Genet 114:1029–1038
Funke C (2008) Hafer, Avena sativa L. Bemühungen um das ehemals wichtigste Futtergetreide. In: Röbbelen G (ed) Die Entwicklung der Pflanzenzüchtung in Deutschland (1908–2008): 100 Jahre GFP eV—eine Dokumentation. Gesellschaft für Pflanzenzüchtung, Göttingen, pp 312–320
Gannibal PB, Klemsdal SS, Levitin MM (2007) AFLP analysis of Russian Alternaria tenuissima populations from wheat kernels and other hosts. Eur J Plant Pathol 119:175–182
Geladi P, Kowalski BR (1986) Partial least-squares regression—a tutorial. Anal Chim Acta 185:1–17
Goloubinoff P, Paabo S, Wilson AC (1993) Evolution of maize inferred from sequence diversity of an Adh2 gene segment from archaeological specimens. Proc Natl Acad Sci USA 90:1997–2001
Groh S, Zacharias A, Kianian SF, Penner GA, Chong J, Rines HW, Phillips RL (2001) Comparative AFLP mapping in two hexaploid oat populations. Theor Appl Genet 102:876–884
Holland JB, Bjornstad A, Frey KJ, Gullord M, Wesenberg DM, Buraas T (2000) Recurrent selection in oat for adaptation to diverse environments. Euphytica 113:195–205
Holland JB, Bjornstad A, Frey KJ, Gullord M, Wesenberg DM (2002) Recurrent selection for broad adaptation affects stability of oat. Euphytica 126:265–274
Jordbruksstatistisk årsbok (Swedish Agricultural Statistics Yearbook) (2000) Statistiska centralbyrån (SCB), Stockholm
Leino MW (2011) Återupptäckt av en historisk havresamling (Rediscovery of a historical oat seed collection). Sven Bot Tidskr 105(1):1–4
Leino MW, Hagenblad J, Edqvist J, Strese EMK (2009) DNA preservation and utility of a historic seed collection. Seed Sci Res 19:125–135
MacKey J (1994) The history of cereal yield increase. Melhoramento 33:37–53
Mattsson B (1997) Svensk växtförädling av havre (Swedish breeding of oats). In: Olsson G (ed) Den svenska växtförädlingens historia (The history of plant breeding in Sweden). The Swedish Academy of Agricultural and Forest Sciences, Stockholm, pp 205–214
Moore-Colyer RJ (1995) Oats and oat production in history and pre-history. In: Welch RD (ed) The oat crop: production and utilization. Chapman & Hall, Landon, pp 1–34
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci USA 70:3321–3323
Nersting LG, Andersen SB, von Bothmer R, Gullord M, Jorgensen RB (2006) Morphological and molecular diversity of Nordic oat through one hundred years of breeding. Euphytica 150:327–337
Olsson G (1997) Gamla lantsorter—utnyttjande och bevarande (Old landraces—utilization and conservation). In: Olsson G (ed) Den svenska växtförädlingens historia (The History of Pant Breeding in Sweden). The Swedish Academy of Agricultural and Forest Sciences, Stockholm, pp 121–130
Palmer SA, Moore JD, Clapham AJ, Rose P, Allaby RG (2009) Archaeogenetic evidence of ancient nubian barley evolution from six to two-row indicates local adaptation. Plos One 4:e6301
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Perrier X, Jacquemoud-Collet JP (2006) DARwin software. http://darwin.cirad.fr/darwin
Reif JC, Zhang P, Dreisigacker S, Warburton ML, van Ginkel M, Hoisington D, Bohn M, Melchinger AE (2005) Wheat genetic diversity trends during domestication and breeding. Theor Appl Genet 110:859–864
Rines H, Molnar S, Tinker N, Phillips R (2006) Oat. In: Kole C (ed) Genome mapping and molecular breeding in plants, cereals and millets. Springer, Berlin, pp 211–242
Schuelke M (2000) An economic method for the fluorescent labeling of PCR fragments. Nat Biotechnol 18:233–234
Semagn K, Bjornstad A, Skinnes H, Maroy AG, Tarkegne Y, William M (2006) Distribution of DArT, AFLP, and SSR markers in a genetic linkage map of a doubled-haploid hexaploid wheat population. Genome 49:545–555
Shannon CE (1948) A mathematical theory of communication. At&T Tech J 27:379–423 and 623–656
Souza E, Sorrells ME (1989) Pedigree analysis of North-American oat cultivars released from 1951 to 1985. Crop Sci 29:595–601
Tenaillon MI, Sawkins MC, Long AD, Gaut RL, Doebley JF, Gaut BS (2001) Patterns of DNA sequence polymorphism along chromosome 1 of maize (Zea mays ssp mays L.). Proc Natl Acad Sci USA 98:9161–9166
Tinker NA, Deyl JK (2005) A curated internet database of oat pedigrees. Crop Sci 45:2269–2272
Tinker NA, Kilian A, Wight CP, Heller-Uszynska K, Wenzl P, Rines HW, Bjornstad A, Howarth CJ, Jannink JL, Anderson JM, Rossnagel BG, Stuthman DD, Sorrells ME, Jackson EW, Tuvesson S, Kolb FL, Olsson O, Federizzi LC, Carson ML, Ohm HW, Molnar SJ, Scoles GJ, Eckstein PE, Bonman JM, Ceplitis A, Langdon T (2009) New DArT markers for oat provide enhanced map coverage and global germplasm characterization. BMC Genomics 10:39
van de Wouw M, Kik C, van Hintum T, van Treuren R, Visser B (2010a) Genetic erosion in crops: concept, research results and challenges. Plant Genet Resour-C 8:1–15
van de Wouw M, van Hintum T, Kik C, van Treuren R, Visser B (2010b) Genetic diversity trends in twentieth century crop cultivars: a meta analysis. Theor Appl Genet 120:1241–1252
van de Wouw M, van Treuren R, van Hintum T (2011) Authenticity of old cultivars in genebank collections: a case study on Lettuce. Crop Sci 51:736–746
Acknowledgments
This work was supported by grants from the Norwegian Research Council, Project number 178273, ‘Safe grains: Mycotoxin prevention through resistant wheat and oats’. We are very grateful to Prof. H. Martens, Norwegian University of Life Sciences, for his kind help on PCA optimization, and to the anonymous referees for helpful and critical comments.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Graner.
This paper is dedicated to Professor emeritus, Dr. Agric. Knut Aastveit, Chair and Professor of Plant Breeding in the Agricultural University of Norway 1968–1988 and a leading force in Nordic plant breeding, on the occasion of his 90th birthday December 12th, 2011.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
He, X., Bjørnstad, Å. Diversity of North European oat analyzed by SSR, AFLP and DArT markers. Theor Appl Genet 125, 57–70 (2012). https://doi.org/10.1007/s00122-012-1816-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-012-1816-8