Abstract
This study concerns spatial genetic patterning, seed flow and the impact of modern varieties in maize populations in Chimaltenango, Guatemala. It uses a collection of 79 maize seed samples from farmers in the area and five samples derived from modern varieties. Bulked SSR markers employed with bulked samples (ten plants) were used. Genetic distances between populations based on these SSR data were used as a measure of co-ancestry. The study describes the genetic variation in space, assesses the association of maize diversity with spatial and environmental descriptors and quantitative traits, and provides a test of the impact of improved varieties. Maize diversity showed significant isolation-by-distance locally, but not regionally. This was interpreted as evidence for a difference between local and regional mechanisms of seed exchange; regional exchange is more related to innovation. There was also a significant association with altitude and ear/grain characteristics (related to racial classifications). Also, consistent evidence for the influence of modern varieties of maize was found, although its impact was limited spatially. It is argued that the spatial distributions of maize diversity are important to consider for germplasm collection, but should be seen as a recent outcome of dynamic processes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Spatial analysis of the genetic structure of crop populations in traditional agricultural systems may yield important insights for their genetic management. (Greene et al. 2002; Guarino et al. 2002). In conservation ecology, ‘landscape genetics’ is the study of fine-scale genetic distributions and their association with environmental features in the landscape (Manel et al. 2003). Such studies contribute to insights in the underlying processes (gene flow, selection) and genetic management requirements (spatial sampling, conservation units).
For crops, spatial approaches might prove crucial in supporting in situ genetic management of populations (crop improvement and biodiversity conservation). In situ genetic management of crops has become more important in the form of participatory or collaborative crop improvement (involving the perceptions and skills of farmers) and in situ conservation of crop diversity (Almekinders and De Boef 2000; Almekinders and Elings 2001; Brush 2004; Cleveland and Soleri 2002; De Boef et al. 1993). Most of these efforts have been local in extent, and upscaling has been indicated as a crucial next step (Smith and Weltzien 2000; Visser and Jarvis 2000). Therefore, it will be important to understand the current crop diversity situation from a multi-scale perspective (Zimmerer 2003).
This study considers local and regional patterns of genetic diversity and focuses on maize (Zea maysL. ssp. mays) in an area of the western highlands of Guatemala. For this crop and area, previous studies have developed insights and hypotheses about seed exchange (Van Etten 2006a, 2006b; Van Etten and De Bruin, in press). The present chapter evaluates part of these hypotheses using genetic data.
The research reported here focused on three research questions. The first question is whether maize populations are genetically structured in space (including altitude). Our previous studies have found that regional seed exchange is relatively low in the area (Van Etten and De Bruin, in press). Previous studies have investigated the spatial structure of maize populations using neutral markers and found little spatial differentiation (Labate et al. 2003; Pressoir and Berthaud 2004; Perales et al. 2005). These findings will be contrasted with the results of this study.
The second question is about which role phenotypic differences play in seed exchange. Phenotypic differences play a role in environmental adaptation, and they may show evidence of farmer preferences in cultivar selection. In previous work we report several, mainly crop-related, motivations for seed introduction and cultivar replacement: to decrease plant height, to increase yield, to decrease the growing cycle, and to improve the grain quality or change its characteristics (Van Etten and De Bruin, in press). On the other hand, differences in ecological adaptation may constrain seed exchange. In the current study, it was attempted to investigate these factors impinging on seed exchange using quantitative trait data. The study will quantify their relative contribution in shaping the geography of gene flow for the whole study area.
The third question is whether modern varieties or derived materials can be found in the area. Modern varieties tend to be different in quantitative traits from farmer materials and measurements of these traits might reveal, which farmer materials derive from modern varieties. However, under farmer conditions, modern varieties change due to admixture and/or selection (Morris et al. 1999; Van Etten and De Bruin, in press). Thus, this study takes a more general approach and investigates whether farmer materials genetically close to improved varieties are also similar to them in plant-related quantitative traits.
Materials and methods
Research area
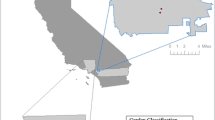
Seed lots were collected from farmers in thirteen communities (caseríos, aldeas) pertaining to four townships (municipios) in Chimaltenango, Guatemala (Table 1). This area represents altitudinal differences between 1500 and 2600 masl (Fig. 2). Most seeds are being recycled by farmers and derive from the previous harvest and from family or neighbours, while a few seed lots come from regional sources (Van Etten and De Bruin in press). Also modern varieties have been introduced into this area in the past, especially since the execution of the Generation and Transfer of Agricultural Technology and Seed Production Project (PROGETTAPS). This was a major project, of national scope, which started in 1986 and ended in the 1990s (Reyes Hernández 1993; Reyes Hernández and García Raymundo 1990; Saín and Martínez 1999). In Chimaltenango the project promoted the adoption of open-pollinated varieties produced by ICTA, in particular V-301 (white kernel), V-302 (yellow kernel), and V-304 (yellow kernel). The first two are adapted to the climatic conditions of the lower part of highland Chimaltenango (1,500–1,900 masl), the last to the higher Central Valley (1,900–2,100 masl). All these varieties are shorter in height and earlier than local cultivars as a result of selection by professional breeders and clearly contrast with native farmer materials in the area with regard to these characteristics (Fuentes 1997). Adoption of these varieties was more frequent in the lower areas (Reyes Hernández 1993; Reyes Hernández and García Raymundo 1990). Agricultural input shops and co-operatives continue to sell seeds of improved varieties (mostly uncertified).
Plant materials
Eighty samples were acquired from randomly selected households among the thirteen communities studied (Table 1). From each household in the sample seed from the seed lot most important for that household was requested. For each seed lot, the location (X, Y, Z) of the household was recorded with a handheld GPS. For each seed lot the following questions were asked: cultivar name, length of time present in household, immediate source, original source (if different), and various agronomic variables. In addition to these farmer materials, five modern varieties developed by ICTA, Guatemala’s national agricultural research institute, were sampled from seeds in stock in ICTA’s seed bank and included in the analysis (Table 2).
Genetic markers
An analysis of Simple Sequence Repeats (SSR) served to determine an index of co-ancestry for the accessions. SSR are neutral genetic markers that are highly polymorphic and therefore very suited for intraspecific studies. Both individuals and genetic markers were bulked in this study. Simple population dissimilarity measures based on bulked samples have been found useful in evolutionary studies, breeding programmes and genetic resource management (Fu 2000). For maize, the feasibility of bulking markers using SSR markers was explored by Xia et al. (2000), who found a high correlation between bulked and non-bulked genetic distances, and a good correspondence with known pedigrees (see also Warburton et al. 2002).
The analysis was conducted at the ICTA biotechnology laboratory. One accession was unavailable for the DNA analysis (n = 84).
Fresh tissue from a bulk sample of ten plants per accession was ground to a fine powder using liquid nitrogen. The powder was incubated at 65°C during 30 min with 500 μl CTAB buffer and 120 μl N-lauroyl-sarcosine 5%, shaking constantly, followed by two chloroform-isoamyl alcohol extractions.
The liquid phase was incubated at 37°C during 30 min with 30 μl of RNase A 10 mg/ml. DNA was precipitated with 1 ml of absolute ethanol stored at −20°C and incubated at −20°C during 15 min. It was centrifuged at 13,000 g during 10 min and the pellet was washed with ethanol (70%). The pellet was re-dissolved in TE buffer (10 mM Tris-HCl pH of 8.00, 1 mM EDTA). It was stored at 4°C. For the obtained DNA dilutions, DNA concentrations were determined with a spectrophotometer using as a conversion factor A 260 nm 1.0 = 50.0 μg/ml.
SSR primers were selected on basis of their equal annealing temperature (56°C), and their distribution in the genome (bin location). The selected primers are shown in Table 3. PCR amplifications were carried out in a total volume of 50 μl, with 5 μl template DNA, 1 × PCR buffer, 2.5 mM MgCl2, 400 μM dNTPs, 1 μM of each primer, and 2 U Taq DNA polymerase.
The samples were mixed 1:1 with ‘stop mix’ (95% formamide, 1 mg xylene cyanole, 1 mg bromophenol blue, 0.5 M EDTA, distilled water) and underwent vertical electrophoresis (Bio-Rad) in a 5% denaturing polyacrylamide gel and silver staining. The thus amplified DNA fragments were recorded manually in an Excel table, coded as present (1) or absent (0).
Quantitative traits
On the 18th of May, 2004, the 85 accessions were sown in an experimental plot at the ICTA Chimaltenango station at 1776 masl. The plot was divided in four repetitions, which contained five incomplete blocks each. Each block was subdivided in 17 parcels containing one accession each. Each parcel consisted of two rows of five planting holes each planted with four plants (=40 plants per accession). Repetitions and blocks lay across the ploughing direction. Assignment of accessions to blocks and parcels for the four repetitions followed an alpha-lattice design (Patterson and Williams 1976).
In this study, phenotypical traits are used to measure genetic variations influencing seed exchange in space. Yield measurements give an indication of divergent ecological adaptation, plant-related phenotypic differences form important motivations for seed exchange and form the main differences between improved and traditional varieties, while ear characteristics remain relatively stable over space (maize races, cf. Wellhausen et al. 1957) and may positively influence seed exchange (for an overview of the variables see Table 4). The traits included in this study were measured for each accession following IBPGR (1991) definitions. Least square means for each variable were estimated using the REML method in the ‘Mixed’ procedure of SAS 9.1 (SAS Institute Inc. 2003), taking into account the effect of differences between repetitions and blocks within repetitions. For all variables, accessions had significant differences. The least square means were used as an input in subsequent analyses.
Data analysis
Genetic distances
The binary SSR data for each accession (n = 84) served to calculate a matrix of pairwise distances. These were calculated as the number of different bands (e.g. {0, 1} and {1, 0}), using GenAIEx 6 (Peakall and Smouse 2005). This distance measure is equivalent to the simple mismatch coefficient (Kosman and Leonard 2005) and is a Euclidean metric (Huff et al. 1993).
Data visualisation
The genetic distance matrix was used to create an unrooted tree diagram using the neighbour-joining method (Saitou and Nei 1987) in the Drawgram programme of the Phylip 3.65 package (Felsenstein 2005). To gain a further impression of the spatial structure of these data, the geographic clusters of accessions were identified visually in this tree diagram and coded with letters, which were mapped using the GPS data taken with each accession. Spatial structure and the influence of modern varieties were statistically tested without reference to discrete clustering patterns, so no postprocessing of the tree diagram was undertaken.
Multivariate analysis: general aspects
Several parts of the analysis relied on a multivariate data analysis to evaluate the relative importance of spatial structure, differential environmental adaptation, and the role of quantitative crop descriptors. This was done by decomposing SSR-based genetic variation by (partial) constrained ordination. This method was introduced into ecology by Borcard et al. (1992) to decompose variation associated with spatial as opposed to environmental variables. All ordination analyses were done in CANOCO 4.5 (ter Braak and Šmilauer 2002) using redundancy analysis (RDA). Mathematically, RDA is an ordination method related to both principal components analysis (PCA) and multiple regressions. RDA (like PCA) reduces a multidimensional data set to a few dimensions. However, in RDA an additional constraint is added to the reduction of data; the resulting axes have to be linear combinations of a set of explanatory variables. RDA differs from multiple regressions in that the Y is composed of a set of several variables.
To be able to analyse the SSR data using RDA, the genetic distance matrix was subjected to a principal coordinate analysis (PCoA), using GenAIEx 6 (Peakall and Smouse 2005). Also, principal coordinates were constructed for parts of the data (see below). Since the distance measure used is a Euclidian metric, all eigenvalues of the PCo’s were positive. The SSR-based principal coordinates were used as the set of dependent variables in RDA. Since grain colour was not significantly associated with SSR-based principal coordinates (evaluated with RDA, using dummy variables for grain colour) for the whole area and subareas, the analysis was done for all colours together. For the third part of the analysis, which focused on the lower areas only, colour (the dummy variable for white) was significant (P < 0.01) and explained 7.1% of the SSR variation. In this analysis, the dummy variable for white was used as a co variable. All significances were determined with permutations under the reduced model in CANOCO 4.5.
Spatial structure
The first part of the analysis is related to the spatial genetic structure of the maize populations. A spatial correlogram was constructed using the pair wise SSR distances to evaluate the presence and extent of isotropic spatial autocorrelation of selectively-neutral genetic diversity in the sample.
Additionally, a multivariate approach was used to evaluate the relation between the SSR data versus spatial distance and altitude. To account for local spatial structure principal coordinates of neighbourhood matrixes (PCNM) were constructed using SpaceMaker2 (Borcard and Legendre 2004). This programme makes principal coordinates of Euclidean distances among sites after setting the values above a certain distance to a constant value (Borcard and Legendre 2002). This maximum local distance (after which relations are not to be considered local anymore) can be determined using different methods, but the implications of these are not fully understood (Borcard and Legendre 2004). Therefore, two different methods were compared: (1) taking the longest distance in the Delaunay Triangulation (DT), and (2) the Relative Neighbourhood Graph (RNG). The difference in results between these two methods did not affect the main findings. X and Y coordinates were added to the spatial explanatory variables in order to model a plane through the data points, representing global spatial structure. Variables were selected using forward selection in CANOCO with P < 0.05.
SSR-based genetic variation was portioned between the set of spatial variables and altitude following Borcard et al. (1992). In this method, the intersection between altitude and the set of spatial variables is calculated as the difference between the gross effect of the set of selected spatial variables (without covariables) and its ‘pure’ effect (taking altitude as covariable). Theoretically, this difference can be negative (Borcard and Legendre 2002). This analysis was done for the whole study area, and repeated for three subareas (see Fig. 1).
Quantitative traits
The second part of the analysis extended the first part of the analysis to include various set of quantitative descriptor variables, related to the yield, plant morphology/phenology and ear morphology, respectively (Table 1). After a check for normality of the quantitative traits, using Q-Q plots in SPSS (SPSS Inc. 2003), yield was log-transformed. In the RDAs only the PCNM spatial descriptor smade using a truncation value derived from a Delaunay triangulation were used.
To be able to partition variation between many sets of variables, the protocol given by Økland (2003) was followed. First, variables were selected for each set using forward selection (P < 0.05). The ‘pure’ effects of all significant variable sets were determined by running RDAs taking the complementary sets as co variables. Then, RDAs were run for all combinations of two variable sets, taking the complementary sets as co variables. The first-order intersections were calculated by subtracting the corresponding ‘pure’ effects of the two sets included in each RDA run. Second-order intersections were calculated as the outcomes of RDA runs on all possible combinations of three variable sets with two complementary co variable sets, subtracting the four corresponding first-order intersections and the three ‘pure’ effects. Likewise, higher-order intersections can be calculated. The last intersection (of all variable sets) is calculated by subtracting all lower-order intersections from the total variance explained.
Since in the approach used no significance levels for combined effects of variable sets can be calculated, the results were simplified using a heuristic method: ‘pruning’ the intersections smaller than L = total variance explained/total number of intersections (Økland 2003). This was first determined for the highest order partial intersection. If a certain intersection (<L) was excluded, its variation was equally distributed among the corresponding intersections of one order lower. Subsequently, the intersections of this order were pruned, and so on. The ‘pure’ effects were not pruned. The results were summarised in a flow diagram, indicating the relative contribution of each factor and factor combination to the total explained variation. Analyses on subareas and on low and high areas (see below) separately, showed that none of the variable sets apart from space and altitude had a significant ‘pure’ effect.
Incidence of modern varieties
The third part of the analysis focused on the possible contribution of improved varieties in the research area on plant characteristics. Modern varieties are different in quantitative variables. Breeding for shorter plants and early flowering have been the main goals of selection (Fuentes 1997). Since farmers reported improved variety names in the lower areas only this analysis focused on the lower part of the study area (communities with average altitude < 2100). The following communities were included: PL, OC, HM, PX, SM, LU, LC, SD (n = 46; see Table 1 and Fig. 2). Principal components based on the SSR-based genetic distance matrix were calculated and used as the dependent variable set in an RDA. If modern varieties have had an impact in the area, it would be expected that farmer cultivars genetically close to these varieties would be more similar in plant-related characteristics. To test this possibility, the following procedure was followed. Forward selection (P < 0.05) between different plant characteristics (Table 4) was undertaken using the SSR-based principal coordinates as response variable. The most significant plant variable resulting from this first step was then used as the response variable in a regression analysis, using the genetic distance to the closest improved variety as the explanatory variable. This second step tests whether farmer seed lots phylogenetically close to improved varieties are also similar phenotypically. This was undertaken for white and yellow materials together, and then separately, i.e. for white cultivars and V-301 (a white variety) and yellow cultivars and V-302 (a yellow variety), respectively.
Results
Germplasm collection
Of the collected seed lots, 37 were white, 40 were yellow, two were black, and one had all three aforementioned kernel colours (pinto). Names of improved varieties were mentioned for seed lots from the lower areas especially, and might serve as a general indication of the possible impact of modern varieties on local germplasm (Van Etten and De Bruin, in press). In at least 12 cases, germplasm originated from farmers in other communities within the same township. Generally, maize was planted within the communities from which they were retrieved. In six cases maize was planted in other communities, in most cases in neighbouring communities, and always in the same township.
Data visualisation
In Fig. 1, the tree diagram for the 84 accessions is presented. From Fig. 1, it is clear that selectively neutral genetic spatial structure exists, as similar two-letter community codes tend to cluster together. The visually determined groups in Fig. 1 contain accessions with a relatively high geographical proximity. Grouping of the accessions is presented geographically in Fig. 2. Most groups are spread over adjacent communities. In two cases similar germplasm is shared between the different subareas. Group H is found in PX (subarea 2) and SD (subarea 3) while group E includes one case outside of its main community, CC (Subarea 2), and is found also in CH (Subarea 1). These cases provide evidence for the existence of some regional gene flow. Groups with high mutual genetic distances (A, B, E, G) are found in high environments (>2,000 masl). This might indicate a lower rate of exchange within communities in these environments, a tendency noted for this area (Van Etten and De Bruin in press).
The improved varieties are clustered close to accessions from low areas, from both subarea 2 and 3. Group F and D (located in subarea 2 and 3, respectively) contain all of the improved varieties. Don Marshall and B-7 cluster together and are close to the root of group F. Also, San Marceño is closer to the root of the group than any farmer cultivars in its branch. V-301 and V-302 are ‘in between’ farmer cultivars in their respective groups, which could be interpreted as support for their influence on the maize gene pool in the area.
Spatial structure
In Fig. 3, a spatial correlogram is presented for the SSR genetic distances of all analysed farmer cultivars (n = 79). This figure shows the degree of isotropic spatial structure over different geographical ranges. The highest degree of correlation is found over small distances, as would be expected in an isolation-by-distance model. Over longer distances (>8 km) a negative correlation is found. This would mean that genetic similarity increases with geographical distance; this has no obvious biological explanation and might be due to the suboptimal structure of the sample for these ranges (confidence statistics refer to the sample, not the entire area). Given the gradual decrease of correlation as distance increases, it can be concluded that over longer distances there is an absence of the isolation-by-distance effect. The turnover point, where the correlation becomes negative, corresponds to the largest inter-sample distance within any subarea (subarea 3 = 8 km). Thus, it might be concluded that isotropic spatial structure is absent between the different subareas (but not within them).
In Tables 5a and b the results of the RDA analysis of the genetic structure of maize populations are presented for the whole area and the three subareas for the two methods employed (DT and RNG). The two methods imply different truncation values which constitute the maximum distance which is still considered as local. The RNG-method produces shorter truncation values for a given collection of points in space than the DT-method. Using a shorter truncation distance in the construction of PCNM spatial descriptors implies that finer spatial structures will receive weight in the statistical analysis.
Irrespective of the method followed (DT or RNG), both space and altitude gives a significant, unique contribution to the structure of maize populations in the redundancy analysis results. For the whole study area, reducing the truncation distance from 31 to 22 km did not improve the overall explained variation much (0.9%). The RNG-based spatial descriptors (Table 5b) only took over some of the variation explained by altitude in the DT-based method (Table 5a). In all subareas, significant spatial structure was demonstrated. In the subareas, the RNG-based spatial descriptors improved the explained variation substantively. This indicates that for the extent of the three subareas (with maximum distances of 4, 7 and 8 km in subarea 1, 2 and 3, respectively), fine, local structures exists. In the relatively flat subareas 1 and 3, no influence of altitudinal differences was noted. However, in subarea 2, which stretches out over a gradient, altitude explained a substantial portion of the variation. However, this could not be distinguished from fine local spatial structure indicated by the RNG-based spatial descriptors.
Quantitative traits
In the redundancy analysis of the SSR-based co-ancestry data (response) versus the quantitative traits, ear characteristics and yield gave significant results, while the set with plant-related characteristics did not show a significant association with the SSR data. Combining the significant qualitative traits (ear characteristics, yield) with the altitude and spatial descriptors (using Delaunay Triangulation, see Table 5a), in total 43.8% of the genetic variation could be explained. Variation was partitioned over pure effects and intersections and all intersections with a value lower than L = 43.8/15 = 2.92 were removed. The largest removed intersection was sized at 1.9 percent; one intersection had a small, negative value (-0.3). Two partial intersections between variable sets remained after simplification of the results: the first-order intersection between ear characteristics and spatial descriptors, and the second-order intersection between yield, spatial descriptors and altitude.
The relative contributions of each factor to the total explained variation are represented in Fig. 4. Spatial descriptors and altitude each have a major share in the total explained variation and their contribution partly overlaps (8.4%). This overlap corresponds to yield (an indicator of environmental adaptation). Yield also gives a small but marginally significant independent contribution (3.8%; P < 0.1). The ear characteristics also relate to an important share in the co-ancestry data. Much of this variation is patterned in space, but ear characteristics also give an independent contribution (9.2%; P < 0.1).
Factors related to the SSR-based genetic diversity of maize in the whole study area. Percentages add up to 100, and represent portions of the total ‘explained’ variation (43.8%). Arrows directly pointing from ear characteristics and yield to co-ancestry represent the sum of the pure effect and the intersection with spatial descriptors and/or altitude
Incidence of modern varieties
The analysis of the possible impact of modern varieties on the germplasm collected focused on the lower area only (communities below 2,100) and included grain colour as a covariable (dummy variable for white vs. other colours). After forward selection on the plant descriptor variable set (Table 4) only the variable remaining was number of leaves. This variable explained 8.6 % of the variation (P < 0.001). However, the other plant-related variables were also significantly associated with genetic diversity (P < 0.05), and correlated with number of leaves. Regression analysis was used to test whether these genetic differences indeed indicated an influence of improved varieties. In Fig. 5, the number of leaves of plants was related to the distance to the closest improved variety. This relationship is significant (P < 0.001), a strong indicator for the influence of improved germplasm on the collected materials. The constant of the equation of the fitted line is 22.8 ± 0.8 (95% confidence interval). The number of leaves of the ICTA varieties V-301 and V-302 fall within this confidence interval (Table 6).
Additional regression analyses evaluated the relation between number of leaves and the distance to V-301 and V-302 only, and to V-301 and V-302 separately for white and yellow cultivars. respectively. All evaluated relationships showed a positive correlation between number of leaves and distance from improved germplasm, as expected. All correlations were significant (P < 0.05), except for the white varieties and V-301 (P < 0.11), which was also the smallest group.
Discussion
Spatial structure
Genetic distances and geographical distances correlate over distances smaller than the maximum extent of the subareas in this study. This finding points to isolation-by-distance causing local spatial structure, presumably the decreasing intensity of seed exchange over growing distances. A previous study in the same area shows that neighbours tend to exchange more seeds with each other than with other community members, community members tend to exchange more seeds with each other than with members of other communities, and also in township-sized areas some containment exists (Van Etten and De Bruin in press). The current study shows over distances greater than those contained within subareas, isolation-by-distance patterns break down, but spatial structure continues to exist. The importance of the X and Y coordinates in the RDA demonstrate that there are clear regional differences in the genetic composition of maize population. This suggests that regionally mutual distances do no longer form the main factor of influence on seed exchange, but that space still structures seed movement in other ways.
These findings can be compared with those of similar studies on maize that used neutral markers. In a study on historical Corn Belt cultivars, Labate et al. (2003) found that genetic distances based on SSR markers did not associate with geographical distances, using a Mantel test of matrix association. The spatial correlogram used in this study is an equivalent to the Mantel test, as it tests isotropic spatial structure. The present study also found no isotropic spatial structure regionally (distances >8 km), but demonstrated it is present locally. Also, by expanding the methods to include non-isotropic spatial structure, it demonstrated regional spatial structure was present.
Using SSR markers, Pressoir and Berthaud (2004) investigated maize from the Central Valleys of Oaxaca collected from communities (longest distance ∼100 km) and found small but significant differentiation levels (FST) among populations and villages. Also, Perales et al. (2005) concluded from an isozyme analysis that two groups of maize collected from two ethnolinguistic groups in Chiapas (longest distance ∼50 km) were not differentiated (low FST).
In the context of a metapopulation, however, low differentiation does not necessarily imply currently high levels of gene flow, as local bottlenecks after colonisation may reduce FST between populations (Pannell and Charlesworth 2000). Arguably, maize as managed by Mesoamerican farmers is structured as a metapopulation, and local bottlenecks are common (Louette 1999). In Oaxaca and elsewhere in Mesoamerica, seed exchange often involves small quantities of seed (Badstue et al. 2005). On the other hand, the low FST values may indicate intensive gene flow in the past (Slatkin 1987; Templeton 1998). Indeed, studies by Pressoir and Berthaud (2004) on maternally inherited DNA confirm this interpretation. In the current study area, the divergence between communities demonstrated by means of genetic distances has arguably a relatively recent origin, whereas the lack of divergence demonstrated by FST measurements in the Mexican studies has a historically more remote origin. The Mexican populations may show divergence when the methods of the present study would be applied to them.
A recent origin for the demonstrated genetic divergence between maize populations of different villages of highland Guatemala has historical grounds, because many rural settlements were created in the course of the nineteenth and twentieth century (Van Etten 2006a). Even so, the study has been able to demonstrate the effect of the contemporary localised seed exchange, which characterises maize agricultural systems in highland Guatemala (Van Etten and De Bruin in press), and other parts of Mesoamerica, including Mexico.
Quantitative traits
Two additional crop related factors were shown to relate to co-ancestry: ear characteristics and yield. The relevance of ear characteristics indicates that these are a good independent predictor for genetic diversity. Apparently, both observed variables, ear characteristics and SSR markers, give a similar indication of ancestry. That ear characteristics are indicative for ancestry is of course assumed in racial classifications (Wellhausen et al. 1957). The significant ‘pure’ effect of ear characteristics also indicates that seed flow based on preferences related to the morphology of ear and grain (Van Etten and De Bruin in press) might have an important influence on the spatial structure of maize populations. Yield was mainly associated with altitude and space. This indicates that environmental adaptation is an important constraint to seed flow. However, it is also demonstrated that there is an important independent contribution of spatial descriptors to the explanations. This might indicate that some underlying environmental factors and/or social limitations to seed flow as yet unidentified play an important role. Social limitation seems likely, as there is a strong local tendency to isolation-by-distance. The independent influence of altitude (unrelated to yield) is less easy to explain. It would be expected that altitude would correspond to yield (as an indicator of adaptation), and have little additional explanatory power. It seems that yield (expressed in one location in one year) is not a comprehensive measure of adaptation. In future work it may be important to use yield data collected a period of years and in different locations to improve the evaluation of environmental adaptation.
Incidence of modern varieties
Van Etten and De Bruin (in press) describe the process of introduction of maize cultivars from outside the community in order to obtain plants lower in stature with a shorter growing cycle. Plant characteristics were significantly related to the SSR co-ancestry data in the lower area (communities below 2,100 masl), but overlapped with other factors, especially space and ear characteristics (data not shown). This indicates that the impact of modern varieties, where it exists, is spatially structured.
Various findings point to an impact of modern varieties. V-301 and V-302 clustered between farmer materials in the tree diagram. Plant-related variables, under selection by professional plant breeders, related significantly to co-ancestry in the dataset for the lower part of the study area. Accessions closer to improved varieties had fewer leaves, as predicted if the data on plant-related genetic differences are to be explained by modern varieties. Also, V-301 and V-302 had the number of leaves predictable from the data. These were the varieties that were introduced successfully in the low part of the study area during the PROGETTAPS project in the 1980s and 1990s (see above).
Taken together, there is strong evidence for an impact of improved varieties in the area in quantitative characters and selectively neutral diversity. However, no (near) identity matches with modern varieties were found. This might be seen as an indication that recent introductions of modern varieties are relatively rare.
Conclusions
The maize populations from Chimaltenango studied here showed clear spatial structure, corresponding to isolation-by-distance locally and to clinal variation regionally. This finding points to different patterns of seed exchange for different spatial ranges. Locally, the intensity of exchange may be expected a rather regular decay over distance between neighbours and members of other communities. This would lead to the observed pattern. The regional pattern reflects, however, that seed exchange between different townships follows a different logic. Regional seed exchange may consist in saltatory movements, there may be different acceptation in different localities, or certain geographical sources may dominate regionally.
Apparently, different mechanisms are at work at different levels; the two spatial levels involve different types of social relationships. Family and neighbours dominate at the local end of the spectrum. Regional exchange involves relations with traders, shop-keepers, NGO personnel, or vague acquaintances (Van Etten and De Bruin, in press). For the first category spatial proximity is relevant, while for the other category different spatial factors dominate, such as centrality (the provincial market). The innovative focus of regional seed exchange may override the spatial factors, as to the innovator the specific characteristics of the seed will tend to be more important than the place it comes from.
Regionally and locally, there is evidence that specific environmental adaptation constrains seed flows, while regionally ear and grain characteristics may influence decision-making on cultivar introduction. The study also demonstrated the impact of improved varieties on genetic diversity and plant characteristics. Comparisons with results for other areas lead to the conclusion that the currently observed patterns of genetic diversity are of rather recent origin.
This study has several implications for genetic management of crop populations in the highlands of Guatemala. Evidence for social constraints to seed flow was found, even though modern germplasm has been successfully adopted in the past. This implies that improved access to (modern) germplasm and information about its availability is needed. As spatial and environmental factors play an important role in structuring the gene pool, spatial sampling imbalances in germplasm for use in breeding will tend to reduce the genetic basis for improvement. Spatial and altitudinal stratification of the area for collection and inclusion of materials in breeding programmes will be necessary to obtain optimal collections. On the other hand, given the relatively small genetic differences between localities and their recent origins, it may not be warranted to constrain gene flow in the study area to maintain diversity. Collaborative farmer-professional maize breeding may be useful in exploiting broad, representative populations in various locations and to strike a balance between improvement and conservation.
References
Almekinders CJM, De Boef W (2000) Encouraging diversity. The conservation and development of plant genetic resources. Intermediate Technology Publications, London
Almekinders CJM, Elings A (2001) Collaboration of farmers and breeders: participatory crop improvement in perspective. Euphytica 122(3):425–438
Badstue LB, Bellon MR, Berthaud J, Ramírez A, Flores D, Juárez X, Ramírez F (2005) Collective action for the conservation of on-farm genetic diversity in a center of crop diversity: an assessment of the role of traditional farmers’ networks. IFPRI-CAPRi, Washington, DC
Boef W de, Amanor K, Wellard K, Bebbington A (1993) Cultivating knowledge. Genetic diversity, farmer experimentation and crop research. Intermediate Technology Publications, London
Borcard D, Legendre P (2002) All-scale spatial analysis of ecological data by means of principal coordinates of neighbour matrices. Ecological Modelling 153(1):51–68
Borcard D, Legendre P (2004) SpaceMaker2 – Users’ guide. Département de sciences biologiques, Université de Montreal, Montréal www.fas.umontreal.ca/biol/legendre
Borcard D, Legendre P, Drapeau P (1992) Partialling out the spatial component of ecological variation. Ecology 73(3):1045–1055
Braak CJF ter, Šmilauer P (2002) Canoco for Windows Version 4.5. Biometris – Plant Research International and Microcomputer Power, Wageningen and Ithaca
Brush SB (2004) Farmers’ bounty. Locating crop diversity in the contemporary world. Yale University Press, New Haven & London
Cleveland DA, Soleri D (2002) Farmers, scientists and plant breeding: integrating knowledge and practice. CABI, Wallingford
Felsenstein J (2005) PHYLIP (Phylogeny Inference Package) version 3.65, Department of Genome Sciences, University of Washington, Seattle http://evolution.genetics.washington.edu/phylip.html
Fu YB 2000. Effectiveness of bulking procedures in measuring population-pairwise similarity with dominant and co-dominant genetic markers. Theor Appl Genetics 100(8):1284–1289
Fuentes MR (1997) Desarrollo de germoplasma de maíz para el Altiplano de Guatemala. Agronomía Mesoamericana 8(1):8–19
Fuentes MR (2002) El cultivo de maíz en Guatemala. Una guía para su manejo agronómico. ICTA, Guatemala
Fuentes MR n.d. ICTA B-7 (ts). ICTA, Guatemala
Greene SL, Gritsenko M, Vandemark G, Johnson RC (2002) Predicting germplasm differentiation using GIS-derived information. In: Engels JMM et al (eds) Managing plant genetic diversity. IPGRI, Rome, pp 405–412
Guarino L, Jarvis A, Hijmans RJ, Maxted N (2002) Geographic Information Systems (GIS) and the conservation and use of plant genetic resources. In: Engels JMM et al (eds) Managing plant genetic diversity. IPGRI, Rome
Huff DR, Peakall R, Smouse PE (1993) RAPD variation within and among natural populations of outcrossing buffalograss [Buchloë dactyloides (Nutt.) Engelm.]. Theoretical and Applied Genetics 86(8):927–934
Kosman E, Leonard KJ (2005) Similarity coefficients for molecular markers in studies of genetic relationships between individuals for haploid, diploid, and polyploid species. Mol Ecol 14(2):415–424
Labate JA, Lamkey KR, Mitchell SE, Kresovich S, Sullivan H, Smith JSC (2003) Molecular and historical aspects of Corn Belt dent diversity. Crop Science 43(1):80–91
Louette D (1999) Traditional management of seed and genetic diversity: what is a landrace? In: Brush SB (ed) Genes in the field. On-farm conservation of crop diversity, Boca Raton, Rome/Ottowa. pp 109–142
Manel S, Schwartz MK, Luikart G, Taberlet P (2003) Landscape genetics: combining landscape ecology and population genetics. Trends in Ecol Evol 18(4):180–197
Økland RH (2003) Partitioning the variation in a plot-by-species data matrix that is related to n sets of explanatory variables. J Veg Sci 14(5):693–700
Pannell JR, Charlesworth B (2000) Effects of metapopulation processes on measures of genetic diversity. Philos Trans Royal Soc B: Biol Sci 355(1404):1471–2970
Patterson HD, Williams ER (1976) A new class of resolvable incomplete block designs. Biometrika 63(1):83–92
Peakall R, Smouse P (2005) GenAlEx 6: Genetic Analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes (online)
Perales HR, Benz BF, Brush SB (2005) Maize diversity and ethnolinguistic diversity in Chiapas, Mexico. Proceedings of the National Academy of Sciences of the United States of America 102(3):949–954
Pressoir G, Berthaud J (2004) Patterns of population structure in maize landraces from the Central Valleys of Oaxaca in Mexico. Heredity 92(2):88–94
Reyes Hernández M (1993) Adopción de variedades mejoradas de maíz: La experiencia de PROGETTAPS en Chimaltenango, Guatemala en 1987–1988. Tikalia 7(1/2):57–75
Reyes Hernández M, García Raymundo SS (1990) La adopción de la tecnología transferida a través del PROGETTAPS para los cultivos de maíz, frijol arbustivo, trigo, papa y crucíferas: Una evalucatión de los primeros dos años de ejecución del proyecto en Chimaltenango, Guatemala. ICTA (unpublished report), Guatemala
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4(4):406–425
SAS Institute Inc. 2003. SAS 9.1, Cary, NC
Slatkin M (1987) Gene flow and the geographic structure of natural populations. Science (New Series) 236(4803):787–792
Smith M, Weltzien E (2000) Scaling-up in participatory plant breeding. In: Almekinders C, De Boef W (eds) Encouraging diversity. The conservation and development of plant genetic resources. Intermediate Technology Publications, London. pp 208–213
Templeton AR (1998) Nested clade analysis of phylogeographic data: testing hypotheses about gene flow and population history. Mol Ecol 7(4):381–397
van Etten J (2006a) Molding maize: the shaping of a crop diversity landscape in the western highlands of Guatemala. Journal of Historical Geography 32(4):689–711
van Etten J (2006b) Changes in farmers’ knowledge of maize diversity in highland Guatemala, 1927/37-2004. J Ethnobiol Ethnomed 2(12)
van Etten J, de Bruin S. Regional and local maize seed exchange and replacement in the western highlands of Guatemala (In press)
Visser B, Jarvis D (2000) Upscaling appoaches to support on-farm conservation. In: Almekinders C, De Boef W (eds) Encouraging diversity. The conservation and development of plant genetic resources. Intermediate Technology Publications, London, pp 141–145
Xia XC, Warburton ML, Hoisington DA, Bohn M, Frisch M, Melchinger AE (2000) Optimizing automated fingerprinting of maize germplasm using SSR markers. Paper presented at Deutscher Tropentag, Hohenheim
Warburton ML, Xia XC, Crossa J, Franco J, Melchinger A, Frisch M, Bohn M, Hoisington DA (2002) Genetic characterization of CIMMYT inbred maize and open pollinated populations using large scale fingerprinting methods. Crop Sci 42(6):1832–1840
Zimmerer KS (2003) Geographies of seed networks for food plants (potato, ulluco) and approaches to agrobiodiversity conservation in the Andean countries. Soc Natural Res 16(7):583–601
Acknowledgements
Member organizations of Seker (Chimaltenango) assisted in the collection and are gratefully acknowledged. Prof. C.J.F. ter Braak of WUR is gratefully acknowledged for his advice on the multivariate analysis. (The usual caveat applies.) The SSR analysis was executed as a component of the project “Molecular characterization of the national maize collection using simple sequence repeats (SSR)” (FODECYT 28-03), financed by the Fondo Nacional de Ciencia y Tecnología (FONACYT).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License ( https://creativecommons.org/licenses/by-nc/2.0 ), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
van Etten, J., Fuentes López, M.R., Molina Monterroso, L.G. et al. Genetic diversity of maize (Zea mays L. ssp. mays) in communities of the western highlands of Guatemala: geographical patterns and processes. Genet Resour Crop Evol 55, 303–317 (2008). https://doi.org/10.1007/s10722-007-9235-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-007-9235-4