Abstract
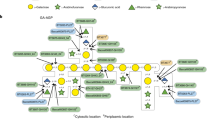
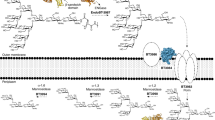
Based on the previous demonstration of surface (S-) layer protein glycosylation in Lactobacillus buchneri 41021/251 and because of general advantages of lactic acid bacteria for applied research, protein glycosylation in this bacterial species was investigated in detail. The cell surface of L. buchneri CD034 is completely covered with an oblique 2D crystalline array (lattice parameters, a = 5.9 nm; b = 6.2 nm; γ ~ 77°) formed by self-assembly of the S-layer protein SlpB. Biochemical and mass spectrometric analyses revealed that SlpB is the most abundant protein and that it is O-glycosylated at four serine residues within the sequence S152-A-S154-S155-A-S157 with, on average, seven Glc(α1-6) residues, each. Subcellular fractionation of strain CD034 indicated a sequential order of SlpB export and glucosylation as evidenced by lack of glucosylation of cytosolic SlpB. Protein glycosylation analysis was extended to strain L. buchneri NRRL B-30929 where an analogous glucosylation scenario could be detected, with the S-layer glycoprotein SlpN containing an O-glycosylation motif identical to that of SlpB. This corroborates previous data on S-layer protein glucosylation of strain 41021/251 and let us propose a species-wide S-layer protein O-glucosylation in L. buchneri targeted at the sequence motif S-A-S-S-A-S. Search of the L. buchneri genomes for the said glucosylation motif revealed one further ORF, encoding the putative glycosyl‐hydrolase LbGH25B and LbGH25N in L. buchneri CD034 and NRRL B-30929, respectively, for which we have indications of a glycosylation comparable to that of the S-layer proteins. These findings demonstrate the presence of a distinct protein O-glucosylation system in Gram-positive and beneficial microbes.








Similar content being viewed by others
Abbreviations
- SlpB:
-
S-layer protein of L. buchneri CD034
- SlpN:
-
S-layer protein of L. buchneri NRRL B-30929
- LbGH25B:
-
Glycosyl‐hydrolase of L. buchneri CD034
- LbGH25N:
-
Glycosyl‐hydrolase of L. buchneri NRRL B-30929
References
Varki, A., Cummings, R.D., Esko, J.D., Freeze, H.H., Stanley, P., Bertozzi, C.R., Hart, G.W., Etzler, M.E.: Essentials of glycobiology, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Habor (2009). 484 pages
Upreti, R.K., Kumar, M., Shankar, V.: Bacterial glycoproteins: functions, biosynthesis and applications. Proteomics 3, 363–379 (2003)
Schmidt, M.A., Riley, L.W., Benz, I.: Sweet new world: glycoproteins in bacterial pathogens. Trends Microbiol. 11, 554–561 (2003)
Hitchen, P.G., Dell, A.: Bacterial glycoproteomics. Microbiology 152, 1575–1580 (2006)
Nothaft, H., Szymanski, C.M.: Protein glycosylation in bacteria: sweeter than ever. Nat. Rev. Microbiol. 8, 765–778 (2010)
Messner, P.: Bacterial glycoproteins. Glycoconj. J. 14, 3–11 (1997)
Messner, P., Sleytr, U.B.: Bacterial surface layer glycoproteins. Glycobiology 1, 545–551 (1991)
Ristl, R., Steiner, K., Zarschler, K., Zayni, S., Messner, P., Schäffer, C.: The S-Layer glycome—adding to the sugar coat of bacteria. Int. J. Microbiol. 2011, 127870 (2011)
Messner, P., Steiner, K., Zarschler, K., Schäffer, C.: S-layer nanoglycobiology of bacteria. Carbohydr. Res. 343, 1934–1951 (2008)
Hartley, M.D., Morrison, M.J., Aas, F.E., Børud, B., Koomey, M., Imperiali, B.: Biochemical characterization of the O-linked glycosylation pathway in Neisseria gonorrhoeae responsible for biosynthesis of protein glycans containing N, N′-diacetylbacillosamine. Biochemistry 50, 4936–4948 (2011)
Logan, S.M.: Flagellar glycosylation—a new component of the motility repertoire? Microbiology 152, 1249–1262 (2006)
Dobos, K.M., Swiderek, K., Khoo, K.-H., Brennann, P.J., Belisle, J.T.: Evidence for glycosylation sites on the 45-kilodalton glycoprotein of Mycobacterium tuberculosis. Infect. Immun. 63, 2846–2853 (1995)
Karlyshev, A.V., Everest, P., Linton, D., Cawthraw, S., Newell, D.G., Wren, B.W.: The Campylobacter jejuni general glycosylation system is important for attachment to human epithelial cells and in the colonization of chicks. Microbiology 150, 1957–1964 (2004)
Szymanski, C.M., Wren, B.W.: Protein glycosylation in bacterial mucosal pathogens. Nat. Rev. Microbiol. 3, 225–237 (2005)
Szymanski, C.M., Yao, R., Ewing, C.P., Trust, T.J., Guerry, P.: Evidence for a system of general protein glycosylation in Campylobacter jejuni. Mol. Microbiol. 32, 1022–1030 (1999)
Wacker, M., Linton, D., Hitchen, P.G., Nita-Lazar, M., Haslam, S.M., North, J.S., Panico, M., Morris, H.R., Dell, A., Wren, B.W., Aebi, M.: N-linked glycosylation in Campylobacter jejuni and its functional transfer into E. coli. Science 298, 1790–1793 (2002)
Ku, S.C., Schulz, B.L., Power, P.M., Jennings, M.P.: The pilin O-glycosylation pathway of pathogenic Neisseria is a general system that glycosylates AniA, an outer membrane nitrite reductase. Biochem. Biophys. Res. Comm. 378, 84–89 (2009)
Fletcher, C.M., Coyne, M.J., Villa, O.F., Chatzidaki-Livanis, M., Comstock, L.E.: A general O-glycosylation system important to the physiology of a major human intestinal symbiont. Cell 137, 321–331 (2009)
Posch, G., Pabst, M., Brecker, L., Altmann, F., Messner, P., Schäffer, C.: Characterization and scope of S-layer protein O-glycosylation in Tannerella forsythia. J. Biol. Chem. 286, 38714–38724 (2011)
Giraffa, G., Chanishvili, N., Widyastuti, Y.: Importance of lactobacilli in food and feed biotechnology. Res. Microbiol. 161, 480–487 (2010)
Helanto, M., Kiviharju, K., Leisola, M., Nyyssölä, A.: Metabolic engineering of Lactobacillus plantarum for production of L-ribulose. Appl. Environ. Microbiol. 73, 7083–7091 (2007)
Peterbauer, C., Maischberger, T., Haltrich, D.: Food-grade gene expression in lactic acid bacteria. Biotechnol. J. 6, 1147–1161 (2011)
Konings, W.N., Kok, J., Kuipers, O.P., Poolman, B.: Lactic acid bacteria: the bugs of the new millennium. Curr. Opin. Microbiol. 3, 276–282 (2000)
Solá, R.J., Griebenow, K.: Glycosylation of therapeutic proteins. BioDrugs 24, 9–21 (2010)
Wells, J.M.: Immunomodulatory mechanisms of lactobacilli. Microb. Cell Fact. 10, S17 (2011)
Möschl, A., Schäffer, C., Sleytr, U.B., Messner, P., Christian, R., Schulz, G.: Characterization of the S-layer glycoproteins of two lactobacilli. In: Beveridge, T.J., Koval, S.F. (eds.) Advances in bacterial paracrystalline surface layers, vol. 252, pp. 281–284. Plenum Press, New York (1993)
Mozes, N., Lortal, S.: X-ray photoelectron spectroscopy and biochemical analysis of the surface of Lactobacillus helveticus ATCC 12046. Microbiology 141, 11–19 (1995)
Konstantinov, S.R., Smidt, H., de Vos, W.M., Bruijns, S.C.M., Singh, S.K., Valence, F., Molle, D., Lortal, S., Altermann, E., Klaenhammer, T.R., van Kooyk, Y.: S layer protein A of Lactobacillus acidophilus NCFM regulates immature dendritic cell and T cell functions. Proc. Natl. Acad. Sci. U. S. A. 105, 19474–19479 (2008)
Mobili, P., los Ángeles Serradell, M., Trejo, S.A., Avilés Puigvert, F.X., Abraham, A.G., Antoni, G.L.: Heterogeneity of S-layer proteins from aggregating and non-aggregating Lactobacillus kefir strains. Antonie van Leeuwenhoek 95, 363–372 (2009)
Messner, P., Schäffer, C., Egelseer, E., Sleytr, U.: Occurrence, structure, chemistry, genetics, morphogenesis, and functions of S-Layers. In: König, H., Claus, H., Varma, A. (eds.) Prokaryotic cell wall compounds—structure and biochemistry, pp. 53–109. Springer, Berlin (2010)
Kawamura, T., Shockman, G.D.: Purification and some properties of the endogenous, autolytic N-acetylmuramoylhydrolase of Streptococcus faecium, a bacterial glycoenzyme. J. Biol. Chem. 258, 9514–9521 (1983)
Fredriksen, L., Mathiesen, G., Moen, A., Bron, P.A., Kleerebezem, M., Eijsink, V.G.H., Egge-Jacobsen, W.: The major autolysin Acm2 from Lactobacillus plantarum undergoes cytoplasmic O-glycosylation. J. Bacteriol. 194, 325–333 (2011)
Rolain, T., Bernard, E., Beaussart, A., Degand, H., Courtin, P., Egge-Jacobsen, W., Bron, P.A., Morsomme, P., Kleerebezem, M., Chapot-Chartier, M.P., Dufrene, Y.F., Hols, P.: O-glycosylation as a novel control mechanism of peptidoglycan hydrolase activity. J. Biol. Chem. 288, 22233–22247 (2013)
Lebeer, S., Claes, I.J.J., Balog, C.I.A., Schoofs, G., Verhoeven, T.L.A., Nys, K., von Ossowski, I., de Vos, W.M., Tytgat, H.L.P., Agostinis, P., Palva, A., Van Damme, E.J.M., Deelder, A.M., de Keersmaecker, S.C.J., Wuhrer, M., Vanderleyden, J.: The major secreted protein Msp1/p75 is O-glycosylated in Lactobacillus rhamnosus GG. Microb. Cell Fact. 11, 15 (2012)
Stepper, J., Shastri, S., Loo, T.S., Preston, J.C., Novak, P., Man, P., Moore, C.H., Havlíček, V., Patchett, M.L., Norris, G.E.: Cysteine S-glycosylation, a new post-translational modification found in glycopeptide bacteriocins. FEBS Lett. 585, 645–650 (2011)
Venugopal, H., Edwards, P.J.B., Schwalbe, M., Claridge, J.K., Libich, D.S., Stepper, J., Loo, T., Patchett, M.L., Norris, G.E., Pascal, S.M.: Structural, dynamic, and chemical characterization of a novel S-glycosylated bacteriocin. Biochemistry 50, 2748–2755 (2011)
Lebeer, S., Verhoeven, T.L.A., Francius, G., Schoofs, G., Lambrichts, I., Dufrene, Y., Vanderleyden, J., De Keersmaecker, S.C.J.: Identification of a gene cluster for the biosynthesis of a long, galactose-rich exopolysaccharide in Lactobacillus rhamnosus GG and functional analysis of the priming glycosyltransferase. Appl. Environ. Microbiol. 75, 3554–3563 (2009)
Denou, E., Pridmore, R.D., Berger, B., Panoff, J.M., Arigoni, F., Brussow, H.: Identification of genes associated with the long-gut-persistence phenotype of the probiotic Lactobacillus johnsonii strain NCC533 using a combination of genomics and transcriptome analysis. J. Bacteriol. 190, 3161–3168 (2008)
Marco, M.L., de Vries, M.C., Wels, M., Molenaar, D., Mangell, P., Ahrne, S., de Vos, W.M., Vaughan, E.E., Kleerebezem, M.: Convergence in probiotic Lactobacillus gut-adaptive responses in humans and mice. ISME J. 4, 1481–1484 (2010)
Heinl, S., Spath, K., Egger, E., Grabherr, R.: Sequence analysis and characterization of two cryptic plasmids derived from Lactobacillus buchneri CD034. Plasmid 66, 159–168 (2011)
Spath, K., Heinl, S., Egger, E., Grabherr, R.: Lactobacillus plantarum and Lactobacillus buchneri as expression systems: evaluation of different origins of replication for the design of suitable shuttle vectors. Mol. Biotechnol. 52, 40–48 (2012)
Liu, S., Skinner-Nemec, K.A., Leathers, T.D.: Lactobacillus buchneri strain NRRL B-30929 converts a concentrated mixture of xylose and glucose into ethanol and other products. J. Ind. Microbiol. Biotechnol. 35, 75–81 (2008)
Zeng, X.Q., Pan, D.D., Guo, Y.X.: The probiotic properties of Lactobacillus buchneri P2. J. Appl. Microbiol. 108, 2059–2066 (2010)
Radovanovic, R.S., Katic, V.: Influence of lactic acid bacteria isolates on Staphylococcus aureus growth in skimmed milk. Bulg. J. Agric. Sci. 15, 196–203 (2009)
Kõll, P., Mändar, R., Smidt, I., Hütt, P., Truusalu, K., Mikelsaar, R.-H., Shchepetova, J., Krogh-Andersen, K., Marcotte, H., Hammarström, L., Mikelsaar, M.: Screening and evaluation of human intestinal lactobacilli for the development of novel gastrointestinal probiotics. Curr. Microbiol. 61, 560–566 (2010)
Danner, H., Holzer, M., Mayrhuber, E., Braun, R.: Acetic acid increases stability of silage under aerobic conditions. Appl. Environ. Microbiol. 69, 562–567 (2003)
Driehuis, F., Oude Elferink, S.J.W.H., Spoelstra, S.F.: Anaerobic lactic acid degradation during ensilage of whole crop maize inoculated with Lactobacillus buchneri inhibits yeast growth and improves aerobic stability. J. Appl. Microbiol. 87, 583–594 (1999)
Holzer, M., Mayrhuber, E., Danner, H., Braun, R.: The role of Lactobacillus buchneri in forage preservation. Trends Biotechnol. 21, 282–287 (2003)
Kung, L.J., Taylor, C.C., Lynch, M.P., Neylon, J.M.: The effect of treating alfalfa with Lactobacillus buchneri 40788 on silage fermentation, aerobic stability, and nutritive value for lactating dairy cows. J. Dairy Sci. 86, 336–343 (2003)
Oude Elferink, S.J.W.H., Krooneman, J., Gottschal, J.C., Spoelstra, S.F., Faber, F., Driehuis, F.: Anaerobic conversion of lactic acid to acetic acid and 1,2-propanediol by Lactobacillus buchneri. Appl. Environ. Microbiol. 67, 125–132 (2001)
Schmidt, R.J., Kung, L.J.: The effects of Lactobacillus buchneri with or without a homolactic bacterium on the fermentation and aerobic stability of corn silages made at different locations. J. Dairy Sci. 93, 1616–1624 (2010)
Heinl, S., Wibberg, D., Eikmeyer, F., Szczepanowski, R., Blom, J., Linke, B., Goesmann, A., Grabherr, R., Schwab, H., Pühler, A., Schlüter, A.: Insights into the completely annotated genome of Lactobacillus buchneri CD034, a strain isolated from stable grass silage. J. Biotechnol. 161, 153–166 (2012)
Liu, S., Leathers, T.D., Copeland, A., Chertkov, O., Goodwin, L., Mills, D.A.: Complete genome sequence of Lactobacillus buchneri NRRL B-30929, a novel strain from a commercial ethanol plant. J. Bacteriol. 193, 4019–4020 (2011)
Liu, S., Bischoff, K.M., Hughes, S.R., Leathers, T.D., Price, N.P., Qureshi, N., Rich, J.O.: Conversion of biomass hydrolysates and other substrates to ethanol and other chemicals by Lactobacillus buchneri. Lett. Appl. Microbiol. 48, 337–342 (2009)
De Man, J.C., Rogosa, M., Sharpe, M.E.: A medium for the cultivation of lactobacilli. J. Appl. Microbiol. 23, 130–135 (1960)
Sleytr, U.B., Messner, P., Pum, D.: Analysis of crystalline bacterial surface layers by freeze-etching, metal shadowing, negative staining and ultrathin sectioning. Methods Microbiol. 20, 29–60 (1988)
Sekot, G., Posch, G., Oh, Y., Zayni, S., Mayer, H., Pum, D., Messner, P., Hinterdorfer, P., Schäffer, C.: Analysis of the cell surface layer ultrastructure of the oral pathogen Tannerella forsythia. Arch. Microbiol. 196, 525–539 (2012)
Laemmli, U.K.: Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 (1970)
Hart, C., Schulenberg, B., Steinberg, T.H., Leung, W.-Y., Patton, W.F.: Detection of glycoproteins in polyacrylamide gels and on electroblots using Pro-Q Emerald 488 dye, a fluorescent periodate Schiff-base stain. Electrophoresis 24, 588–598 (2003)
Taylor, A.M., Holst, O., Thomas-Oates, J.: Mass spectrometric profiling of O-linked glycans released directly from glycoproteins in gels using in-gel reductive β-elimination. Proteomics 6, 2936–2946 (2006)
Packer, N.H., Lawson, M.A., Jardine, D.R., Redmond, J.W.: A general approach to desalting oligosaccharides released from glycoproteins. Glycoconj. J. 15, 737–747 (1998)
Pabst, M., Altmann, F.: Influence of electrosorption, solvent, temperature, and ion polarity on the performance of LC-ESI-MS using graphitic carbon for acidic oligosaccharides. Anal. Chem. 80, 7534–7542 (2008)
Stadlmann, J., Pabst, M., Kolarich, D., Kunert, R., Altmann, F.: Analysis of immunoglobulin glycosylation by LC-ESI-MS of glycopeptides and oligosaccharides. Proteomics 8, 2858–2871 (2008)
Lee, Y.: High-performance anion-exchange chromatography for carbohydrate analysis. Anal. Biochem. 189, 151–162 (1990)
Pabst, M., Bondili, J.S., Stadlmann, J., Mach, L., Altmann, F.: Mass + retention time = structure: a strategy for the analysis of N-glycans by carbon LC-ESI-MS and its application to fibrin N-glycans. Anal. Chem. 79, 5051–5057 (2007)
Hanisch, F.-G., Müller, S.: Approaches to the O-glycoproteome. In: The proteomics protocols handbook. pp. 439–457. Humana Press Inc., Totowa, New York (2005)
Masuda, K., Kawata, T.: Characterization of a regular array in the wall of Lactobacillus buchneri and its reattachment to the other wall components. J. Gen. Microbiol. 124, 81–90 (1981)
Åvall-Jääskeläinen, S., Palva, A.: Lactobacillus surface layers and their applications. FEMS Microbiol. Rev. 29, 511–529 (2005)
van Roosmalen, M.L., Geukens, N., Jongbloed, J.D.H., Tjalsma, H., Dubois, J.-Y.F., Bron, S., van Dijl, J.M., Anné, J.: Type I signal peptidases of Gram-positive bacteria. Biochim. Biophys. Acta, Mol. Cell Res. 1694, 279–297 (2004)
Hynönen, U., Palva, A.: Lactobacillus surface layer proteins: structure, function and applications. Appl. Microbiol. Biotechnol. 97, 5225–5243 (2013)
Kleerebezem, M., Hols, P., Bernard, E., Rolain, T., Zhou, M., Siezen, R.J., Bron, P.A.: The extracellular biology of the lactobacilli. FEMS Microbiol. Rev. 34, 199–230 (2010)
Steiner, K., Hanreich, A., Kainz, B., Hitchen, P.G., Dell, A., Messner, P., Schäffer, C.: Recombinant glycans on an S-layer self-assembly protein: a new dimension for nanopatterned biomaterials. Small 4, 1728–1740 (2008)
Zarschler, K., Janesch, B., Pabst, M., Altmann, F., Messner, P., Schäffer, C.: Protein tyrosine O-glycosylation—a rather unexplored prokaryotic glycosylation system. Glycobiology 20, 787–798 (2010)
Janesch, B., Messner, P., Schäffer, C.: Are the surface layer homology domains essential for cell surface display and glycosylation of the S-layer protein from Paenibacillus alvei CCM 2051T? J. Bacteriol. 195, 565–575 (2012)
Huard, C., Miranda, G., Wessner, F., Bolotin, A., Hansen, J., Foster, S.J., Chapot-Chartier, M.-P.: Characterization of AcmB, an N-acetylglucosaminidase autolysin from Lactococcus lactis. Microbiology 149, 695–705 (2003)
Claes, I.J.J., Schoofs, G., Regulski, K., Courtin, P., Chapot-Chartier, M.-P., Rolain, T., Hols, P., von Ossowski, I., Reunanen, J., De Vos, M.D., Palva, A., Vanderleyden, J., De Keersmaecker, S.C.J., Lebeer, S.: Genetic and biochemical characterization of the cell wall hydrolase activity of the major secreted protein of Lactobacillus rhamnosus GG. PLoS One 7, e31588 (2012)
Faridmoayer, A., Fentabil, M.A., Mills, D.C., Klassen, J.S., Feldman, M.F.: Functional characterization of bacterial oligosaccharyltransferases involved in O-linked protein glycosylation. J. Bacteriol. 189, 8088–8098 (2007)
Hug, I., Feldman, M.F.: Analogies and homologies in lipopolysaccharide and glycoprotein biosynthesis in bacteria. Glycobiology 21, 138–151 (2010)
Acknowledgments
We thank Siqing Liu (U.S. Department of Agriculture, Agricultural Research Service, Renewable Product Technology Research Unit, University of Illinois, USA) for kindly providing L. buchneri NRRL B-30929, and Andrea Scheberl and Sonja Zayni for excellent technical assistance.
Financial support came from the Austrian Science Fund FWF, projects P21954-B20 (to C.S.) and P24305-B20 (to P.M.), the PhD programme “BioToP - Biomolecular Technology of Proteins” (Austrian Science Fund, FWF project W1224), the Hochschuljubiläumsstiftung der Stadt Wien, project H-2442/2012 (to J.A.), and the Christian Doppler Laboratory for Genetically Engineered Lactic Acid Bacteria (to R.G.).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Anzengruber, J., Pabst, M., Neumann, L. et al. Protein O-glucosylation in Lactobacillus buchneri . Glycoconj J 31, 117–131 (2014). https://doi.org/10.1007/s10719-013-9505-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10719-013-9505-7