Abstract
Selective pressure from parasites is thought to maintain the polymorphism of major histocompatibility complex (MHC) genes. Although a number of studies have shown a relationship between the MHC and parasitic infections, the fitness consequences of such associations are less well documented. In the present paper, we characterised the variation in exon 2 of MHC class II DRB gene in the root vole and examined the effects of that gene on parasite prevalence and winter survival. We identified 18 unique exon 2 sequences, which translated into 10 unique amino acid sequences. Phylogenetic analysis revealed the presence of three distinct clusters, and allele distributions among these individuals suggested that the clusters correspond to three different loci. Although the rate of synonymous substitutions (dS) exceeded the rate of nonsynonymous substitutions (dN) across sequences, implying purifying selection, dN was significantly elevated at antigen-binding sites, suggesting that these sites could be under positive selection. Screening for parasites revealed a moderate prevalence of infection with gastrointestinal parasites (24 % infected), but a high infection rate for blood parasites (56 % infected). Infection with the blood parasite Babesia ssp. decreased survival almost twofold (25.7 vs. 13.9 %). Animals possessing the amino acid sequence AA*08 survived better than others (44.9 vs. 22 %), and they were infected with Babesia ssp. less often (13.9 vs 25.7 %). In contrast, individuals carrying allele AA*05 were infected more often (31.7 vs. 15.3 %). Heterozygosity at one of the putative loci was associated with a lower probability of infection with Babesia ssp., but at the other locus, the association was reversed. The unexpected latter result could be at least partly explained by the increased frequency of the susceptible allele AA*05 among heterozygotes. Overall, we demonstrate that infection with Babesia ssp. is a strong predictor of winter survival and that MHC genes are important predictors of infection status as well as survival in the root vole.
Similar content being viewed by others
Introduction
Genes of the major histocompatibility complex (MHC) are a crucial element of the adaptive immune response of vertebrates (Trowsdale 1995). They code membrane proteins that present antigens to T lymphocytes and thus are involved in mounting an immune response against pathogens. The MHC has been demonstrated to influence parasitic infections across a broad taxonomic range of non-model species in natural populations, and as such, it is believed that host-parasite interactions can drive high levels of polymorphism, which are characteristic of MHC loci (reviewed in Bernatchez and Landry 2003; Piertney and Oliver 2006; Spurgin and Richardson 2010). Three primary mechanisms explaining the role of parasites in maintaining MHC polymorphism have been proposed, although the determination of their relative importance remains an empirical challenge (Spurgin and Richardson 2010). According to the heterozygote advantage hypothesis, heterozygotes can be expected to have higher fitness than homozygotes, as they express more MHC alleles and thus can recognise a wider array of pathogen-derived antigens (Doherty and Zinkernagel 1975; Hughes and Nei 1988). The second mechanism, rare-allele advantage, assumes that parasites are most likely to adapt to the most frequent host genotypes, and thus rare alleles are more often associated with parasite resistance (Bodmer 1972; Potts and Wakeland 1990; Borghans et al. 2004). If the selective pressure from a parasite changes temporally (Hill et al. 1991) or spatially (Kloch et al. 2010; Loiseau et al. 2011), this change may drive the third mechanism, fluctuating selection (Hedrick 2002).
Although associations between MHC genes and infection with parasites are relatively well documented (reviewed in Spurgin and Richardson 2010), their fitness consequences are less well understood. Nevertheless, survival-related MHC alleles have been observed in the Seychelles warbler (Brouwer et al. 2010), Soay sheep (Paterson et al. 1998), and brushtail possum (Banks et al. 2010). Additionally, several other studies have documented this relationship in the context of parasitic infections. For example, a higher survival rate of heterozygotes has been observed in red junglefowl during an outbreak of coccidiosis (Worley et al. 2010), and a study of the Soay sheep revealed lower mortality in juveniles possessing MHC alleles associated with resistance against helminths (Paterson et al. 1998). Among fish species, MHC genotype influenced the survival of embryos that had been exposed to bacterial infection (Wedekind et al. 2004), and individuals with homozygous MHC genotypes survived better when infected with flukes than did heterozygotes (Hedrick et al. 2001). In frogs, alleles from the MHC class IIB locus were associated with increased survival following experimental infection with chytridiomycosis, an emerging disease (Savage and Zamudio 2011). In contrast, although MHC heterozygosity increased survival in male chamois, there was no evidence for an MHC-related survival advantage in populations exposed to scabies (Schaschl et al. 2012).
In the present paper, we characterised the variation in MHC class II DRB gene in the root vole and examined the effects of that gene on parasite prevalence and winter survival in a semi-natural population. The voles are small mammals, commonly used in ecological (e.g. Norrdahl and Korpimäki 2005; Borkowska et al. 2009) and parasitological (e.g. Bajer et al. 2001; Burthe et al. 2008) studies, they were also subject of several studies of MHC polymorphism in the wild (e.g. Oliver et al. 2009; Kloch et al. 2010). During the winter, the costs of pathogen tolerance may increase as a result of higher energy expenditures and lower food availability. Moreover, in small mammals, winter mortality is the major factor shaping population dynamics (Jędrzejewska et al. 2004; Norrdahl and Korpimäki, 2005). Our aims were to determine (i) whether MHC DRB gene is associated with infection by blood and gastro-intestinal parasites and (ii) whether infection with those parasites predicts mortality in the sensitive winter period. Given the significant associations of the MHC with infection and of infection with survival, we also aimed to determine (iii) whether alleles that affect infection are significant predictors of survival.
Methods
Study site and species
We studied a semi-natural population of the root vole (Microtus oeconomus) in Biebrza National Park, the largest natural wetland habitat in Poland. Root vole is small rodent (40–70 g) inhabiting open, wet habitats, such as river banks, swamps, marshes etc. The population density varies between years and throughout the breeding season. In Biebrza National Park, the lowest recorded density was 1 individuals/ha, and the highest exceeded 300 individuals/ha (Borowski 2002). Root voles live up to two years, but in the wild, they rarely survive much longer than one year. The breeding season lasts from April to October, females typically usually have 3 or 4 litters per year, and the average litter size is 2.5 (range 2–9) (Borkowska et al. 2009).
The studied population inhabited a 1 ha plot (100 × 100 m) situated on a sedge mire. The study site was enclosed with a 1 m-high fence made of coated wire that extended 50 cm underground. The fence prevented the voles from migrating and protected them from terrestrial predators such as weasels and polecats (Borowski 2011). The enclosure was set up in the autumn of 2007. To avoid possible effects associated with the existing social structure in the fenced area, we removed all of the resident voles and populated the enclosure with 53 animals caught in the nearby area. The animals were trapped in several locations within about 500 m range from the enclosure. All sites were situated in the same mire habitat, which diminished the risk of transferring new parasite species to the enclosure.
Field experiment and handling of animals
In November 2008, the enclosure was divided into two parts (50 × 100 m each) using a fence. The reason for dividing the enclosure was to investigate the effect of gastro-intestinal nematodes on survival by experimentally treating half of the animals with an anti-nematode drug (see below). However, due to the very low prevalence of nematodes in our population (see the results), this aim could not be achieved. In each part, we set up 66 wooden live traps arranged in a 6 × 11 grid. The traps were checked twice per day: at approximately 8 am and at dusk. If the night-time temperature dropped below 0 °C, the traps were closed for the night. Captured animals were transported to a nearby field station where they were sexed, weighed and placed individually in standard laboratory cages. They were provided with bedding and nest material, and they had unlimited access to water and standard rodent feed supplemented with fresh carrots and apples.
Upon arrival, the newly captured voles were marked by toe-clipping. At most, we removed one toe in each paw, which resulted in permanent, unique patterns allowing for identification of each individual. Although this method is invasive, it is acceptable when tissue samples are required in addition to marking (Gannon et al. 2007). Blood was used for parasite screening and tissue was preserved for molecular analysis. The procedures followed ethical regulations and were accepted by the Local Ethical Committee in Kraków (permission no. 76/2008).
In November 2008, animals from the experimental group were injected subcutaneously with Ivomec (Merck, 0.2 μg/g body mass), an anti-nematode drug widely used in rodent studies (e.g., Scantlebury et al. 2007), and the control group was injected with propylene glycol, an Ivomec medium. The injections were administered within 24–72 h after trapping. Next, the animals were transferred to fresh cages, where they were kept for 2–10 days until no additional voles were captured in the field for 3 consecutive days. All animals were then released back into the enclosure. In January 2009 and March 2009, the animals were recaptured using the same method, and in January 2009 the Ivomec treatment (or propylene glycol in the control group) was repeated. The periods between consecutive trappings were about 60 days.
Overall, in November 2008, we captured 167 animals, comprising 92 individuals in the experimental group (37 females, 55 males), and 75 in the control group (27 females, 48 males). In January 2009, we recaptured 48 animals (20 females, 28 males) and in March 2009, 31 animals (12 females, 19 males).
Parasite screening
We analysed two groups of parasites: gastrointestinal helminths and blood microparasites (bacteria and protozoa). Infection with blood parasites was detected in stained blood smears prepared from the blood collected during toe-clipping upon the arrival of the animals at the laboratory. The smears were air-dried, fixed in absolute methanol and stained for 40 min in 5 % Giemsa solution (Merck). The parasites were identified under 1000× magnification based on their colouration, size and shape and compared with specimens provided by the Dept. of Parasitology, University of Warsaw. The parasites in 100 fields of vision (approx. 100,000 red blood cells) were counted as described previously (Bajer et al. 2001; Kloch et al. 2010).
To investigate the prevalence of helminths, we collected faeces from the cages in which animals were kept before the anthelmintic treatment in November 2008. Gastrointestinal helminth burden was determined based on faecal egg count (FEC) using a modified McMaster technique (Dunn and Keymer 1986). FEC is a non-invasive method widely used for determining helminth infections in ecological studies (reviewed in Sommer 2005). The amount of analysed faeces was adjusted to the small body size of the voles; we used 0.5 g of faeces per individual, collected over a period of three days. Chambers were examined under 100–200× magnification. All helminth eggs were counted and identified following the protocol described by Thienpont et al. (1986).
MHC polymorphism
Primer design
We designed PCR primers to amplify the second exon of MHC DRB class II in the root vole. DRB is typically the most variable one among MHC II genes (Robinson et al. 2003) and it has been commonly used in MHC studies in wild populations (e.g. Harf and Sommer 2005; Froeschke and Sommer 2005; Kloch et al. 2010). The variance at given MHC gene is likely to represents the variation at other MHC genes due to the linkeage disequlibrium between MHC genes (Marsh et al. 2000).
In the first step, using non-specific rodent primers (forward: 5′GAGTTGTCATTTCTACAACG3′ (Smulders et al. 2003), reverse: 5′GGACRCRTWCTGCAGAYACAACTA3′ (designed by the authors based on consensus sequences from bank voles and other rodents)), we amplified the 166 bp part of the second exon MHC DRB coding peptide-binding region of the molecule. The PCR reaction volume was 20 μl, and the mix contained 2 μl of the PCR buffer with (NH4)2SO4 buffer (Fermentas), 2.0 mM MgCl2, 0.2 mM of each dNTP, 0.2 μM of each primer, 0.2 U of Taq polymerase (Fermentas) and 1 μl of the template. The program started with 3 min at 95 °C followed by 35 cycles: 30 s at 94 °C, 30 s at 56 °C, and 90 s at 72 °C, with a final elongation at 72 °C for 3 min. The results were checked using agarose gel electrophoresis and the product was then purified enzymatically using ExoSap (Fermentas) and sequenced in both directions using a BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Bioscences).
The next step was to amplify regions localised outside of the resulting sequence. To do so, we designed primers spanning from the 1st to the 3rd exon based on the newly obtained 166 bp sequences from the root vole and an alignment of mammalian 1st and 3rd exons derived from the GenBank. One pair of the “trans-exon” primers consisted of a forward primer situated in the 1st exon (EV1F 5′TGGCAGCTGTGRTCCTG3′) and a reverse primer localised in the middle of the 2nd exon (MovR1 5′GCCCACATCACTGTCRAAGC3′). The forward primer of the second pair was located in the 2nd exon (Mov F1: 5′GCTTYGACAGTGATGTGGGC3′) and the reverse one in the 3rd exon (EV1R: 5′CAGACCAGGAGGTTGTGRTG3′). Because the introns within the MHC DRB are long, we used cDNA as a template. The RNA was extracted from 6 samples from the spleen or liver, taken from voles used in the pilot study in September 2008. The samples were preserved in RNAlater (Qiagen) and extracted using an RNeasy Mini Kit (Qiagen). The reverse transcription was performed with an Omniscript RT Kit (Qiagen) with Recombinant RNasin® Ribonuclease Inhibitor (Promega) and oligo-dT(N) primer (Sigma). The PCR reaction volume was 20 μl and the mix contained 2 μl of the PCR buffer with (NH4)2SO4 buffer (Fermentas), 1.5 mM of MgCl2, 0.2 mM of each dNTP, 0.4 μM of each primer, 0.1 U of Taq polymerase (Fermentas) and 1.5 of the cDNA template. The initial denaturation lasted 15 min at 94 °C and was followed by 35 cycles: 94 °C for 30 s, 56 °C for 30 s, and 72 °C for 90 s, with a final elongation at 72 °C for 3 min. The product was purified using ExoSAP and sequenced in both directions with a BDT BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems).
In the third step, based on the newly sequenced distal regions of the MHC DRB 2nd exon in the root vole, we designed the primers MioeL 5′-GAGTGTCRTTTYTACAAYGGGAC3′ and MioeR 5′TGCWCAGTGAAGCYMTC3′. These primer sequences were located in conserved regions close to the exon’s edges and amplified 196 bp-long part of the 2nd exon. These primers were used in all further procedures.
Genotyping
As the DRB locus often occurs in several copies in microtine rodents’ genomes, we used direct sequencing with Roche 454, which is best suited to reliably genotyping complex, multilocus systems (Babik et al. 2009; Galan et al. 2010; Kloch et al. 2010; Zagalska-Neubauer et al. 2010). DNA from 167 root voles was isolated using a Nucleospin Tissue Kit (Macherey & Nagel). The MHC DRB was amplified using 454 fusion primers. The forward primer consisted of the MioeL primer, a 6 bp tag and the 454 amplicon A forward primer (5′GCCTCCCTCGCGCCATCAG3′). Similarly, the reverse primer was composed of the MioeR primer, a 6 bp tag and the 454 amplicon A reverse primer 5′GCCTTGCCAGCCCGCTCAG3′). The PCR mix consisted of 7.5 μl of Qiagen Master Mix, 5.7 μl of ddH2O, 0.1 μM of each primer and 1.5 μl of the genomic DNA. The initial denaturation lasted 15 min at 94 °C and was followed by 35 cycles: 94 °C for 30 s, 56 °C for 30 s, and 72 °C for 90 s, with the final elongation at 72 °C for 3 min. The concentration of the PCR product was estimated by eye from agarose gel, and PCR products were pooled into approximately equimolar quantities (Babik et al. 2009). We used 8 different tags attached to F primers and 3 tags attached to R primers, obtaining 24 unique combinations to identify amplicons from individuals. Because the total number of animals was 167, we arranged them into 8 pools and each pool was analysed in a separate lane during 454 sequencing. The pools were purified using the MinElute PCR Purification Kit (QIAGEN) and sequenced in a single 454 FLX run according to the 454 Amplicon Sequencing protocols provided by the manufacturer (Roche 454) at the Functional Genomics Center, Uni/ETH Zurich. Additionally, to investigate whether the DRB sequences found in gDNA are transcribed, we amplified and sequenced both cDNA (coverage 82–111) and genomic DNA (coverage 229–435) from 5 individuals used in the pilot study.
In the first step of genotyping, we excluded individuals with coverage below 80, which resulted in the exclusion of 11 individuals (coverages of 1–19). In 454 sequencing, artefacts generated repeatedly through 1 bp indels in homopolymer tracts are especially common, with approximately a 1 % error rate. As the frequency of a given artefact per amplicon may be considerable (Rozera et al. 2009), we excluded all sequences with indels. The probability of obtaining more than two identical substitution-type artefacts per amplicon is negligibly low (Galan et al. 2010), so we also removed sequences that occurred in a single copy or in only two copies. The method described here may not completely eliminate all artefacts such as PCR chimeras (Galan et al. 2010); therefore, we applied an additional procedure (Zagalska-Neubauer et al. 2010). In each individual, starting from the sequences with the lowest frequency, we visually inspected whether any of them could result from (1) a recombination of more common variants present in the individual or (2) a single substitution in any of the more common variants. We found that 38 of the 41 variants (92.7 %) occurring at a per-individual frequency below 3 % might originate from substitution/recombination, and hence they were considered artefacts. In contrast, no sequences with a frequency between 4 and 8 % (n = 19) per individual could be explained in this way; thus, we could safely assume that variants above a 4 % threshold were genuine.
Analysis of sequences and signatures of selection on MHC
The phylogenetic relationships between MHC sequences were analysed in MEGA4 (Tamura et al. 2007) using a tree constructed with the neighbour-joining method. Using the exact tests in Genepop (Rousset 2008), we assessed whether the alleles in putative DRB loci (forming separate clusters in the gene tree) were in Hardy–Weinberg equilibrium. In LinkDos (Garnier-Gere and Dillmann 1992), we tested for genotypic linkage disequilibrium between alleles among putative loci.
We used two approaches to test for selection processes acting on the MHC. The average rates of synonymous (dS) and nonsynonymous (dN) substitutions were computed using MEGA4 for all sites, and separately for positions that code amino acids forming the antigen-binding sites (ABS) in human HLA DRB (Reche and Reinherz 2003) and positions outside ABS. Another test was performed by fitting three models of codon evolution in PAML (Yang 1997). These were: (i) M0: one ω (dN/dS ratio), (ii) M7: nearly neutral (ω ≤ 1) with the beta distribution approximating ω variation, and (iii) M8: positive selection (a proportion of sites evolving with ω > 1) with the beta distribution approximating ω variation. The goodness-of-fit of the models was evaluated using AIC (Posada and Buckley 2004).
Statistics
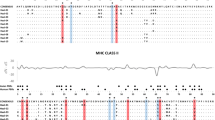
We focused on “amino acid alleles” rather than on “nucleotide alleles”, as some of the nucleotide sequences identified in the present study had the same amino acid composition and we were interested in the effect of the functional, rather than the nucleotide, sequence diversity of MHC molecules. We assigned the amino acid alleles names beginning with “AA*” to avoid confusion with the nucleotide alleles named Mioe-DRB. Allele AA*02 was present in all animals and therefore was not included in the models. We also excluded from the analyses the two rarest alleles, AA*09 and AA*10, as they were only present in 2 and 3 individuals, respectively. The amino acid alleles grouped into 3 distinctive clusters (see Fig. 1), but the cluster I consisted only of the single allele AA*02, so overall, we considered the presences of 6 alleles from two clusters (II and III).
In all analyses of survival, we used generalised linear models with a binary response, binomial error distribution and log link function. Because the animals were re-captured twice, we fitted separate models for survival of the entire study period (from November 2008 to March 2009) and for early (from November 2008 to January 2009) and late winter (from January 2009 March 2009). We analysed two sets of models with either survival or presence of parasites as the response variable. All models included host sex as a categorical variable along with body mass as a continuous covariate.
The low prevalence of nematodes prevented us from detecting the effects of the Ivomec treatment (control vs. injection with Ivomec). Nevertheless, we included the treatment in all models, which allowed us to statistically control for its possible side-effects on survival or infection with other parasites. When analysing the effect of the MHC on survival, we fitted two types of models, the explanatory variable being either the presence of an MHC allele or heterozygosity at a putative MHC locus. Models of the effect of parasites on survival also included as explanatory variables the presence of parasite species that occurred in over 10 % of hosts and first-term interactions between the parasites.
In our models of the effect of the MHC on infections, we quantified the parasitic infection as the presence of a given parasite or—in the case of blood parasites—the intensity of infection. We analysed parasite presence using the binomial error distribution and log link function. In the case of blood parasites, infection intensity was expressed as the number of infected blood cells per 100 fields of vision and fitted using generalised linear models with a negative binomial distribution and logit link. Similarly to the models of survival, the explanatory variables were: treatment, host sex, body mass, and the presence of MHC alleles.
Significance of the terms in the model was determined using deletion testing (Crawley 2007). All analyses were performed with R statistical software, ver. 2.8.1 (R Development Core Team 2008) using the glm or glm.nb functions. To estimate the critical p value for the effect of MHC alleles, we applied a modified false discovery rate procedure (Benjamini and Yekutieli 2001), which is an alternative to the Bonferroni correction and regarded as the best practical solution to the problem of multiple comparisons (Narum 2006).
Results
Prevalence and intensity of infection
We collected coprological samples from 159 animals. Gastrointestinal helminths were found in 24.5 % of voles (n = 39), and most of them were cestodes. Nematodes occurred sporadically: Trichuris was present in 3.7 % of voles (n = 6) and Syphacia in only 1 individual (0.7 %). The prevalence of the two cestode genera, Apostatandria and Rodontolepis, was higher than that of nematodes, both cestodes being found in 10.1 % of voles.
We collected blood smears from 148 animals. Overall, the prevalence of blood parasites was 58.7 %. We identified two protozoa: Babesia ssp. was present in 22.3 % of the individuals (n = 33), and Trypanosoma ssp. in 7.4 % (n = 11). The bacteria Mycoplasma ssp. was present in 43.9 % of individuals (n = 65), and Bartonella ssp. in only 2 individuals (1.3 %). Due to their low prevalence, Bartonella ssp. and Trypanosoma ssp. were excluded from further analyses. On average, we found 3.4, 6.1, and 3.7 infected blood cells per 100 fields of vision for Mycoplasma ssp., Babesia ssp., and Trypanosoma ssp., respectively. Most animals infected with blood parasites carried only one parasite genus (74.5 % of infected hosts, n = 65), two were found in 25 % of hosts (n = 22), and three in only one individual. Assuming random occurrence of parasites, the single parasite infections should be present in 72.3 % of animals, infection with two geni in 26.8 %, with three in 0.9 %, and 0.01 % with four, and the observed values did not differ from expected (χ2 = 0.146, p > 0.985).
MHC polymorphism
Amplification primers were identified in 47,151 reads. After the removal of incomplete or ambiguous sequences, we obtained 45,405 reads, and the mean coverage at this stage was 250.63 reads per amplicon (SD = 76.04, range 83–532). Based on these reads, we identified 18 unique nucleotide sequences in 167 animals. The sequences have been submitted to EMBL-Bank (accession no. HE578727-HE578744). We have confirmed the expression of the following sequences: Mioe-DRB*02, Mioe-DRB*03, Mioe-DRB*05, Mioe-DRB*07, Mioe-DRB*09 and Mioe-DRB*12, which were present in both the genomic DNA and the cDNA of the six individuals investigated. The allele Mioe-DRB*04 was identified as a non-functional allele because it was present in two genomic DNA samples but was not found in cDNA. Consequently, Mioe-DRB*04 was not incorporated into the models.
The mean number of potentially functional alleles per individual was 4.6 (SD = 0.96, range 3–6). Among 17 putative functional alleles, we found 10 unique amino acid sequences and each individual carried, on average, 4.02 amino acid alleles (SD = 0.68, range 3–5). The analysis of similarity between the sequences revealed three distinct clusters (Fig. 1). Because none of the individual voles had more than two alleles belonging to the same cluster and each individual possessed at least one allele from each cluster, the clusters likely represent separate loci. When treated as belonging to separate loci, the alleles in all three clusters were in Hardy–Weinberg equilibrium (Table S1, Supplementary Materials) in accord with their locus status (i.e., some alleles at different loci should be linked, resulting in an excess of false ‘heterozygotes’). The expression of alleles from each putative locus (cluster) has been verified, indicating that all loci were functional. However, we found no expression of the allele Mioe-DRB*04 (cluster III), which suggests that the expression pattern may be haplotype-specific.
Our results suggest that the MHC DRB sequences were under purifying selection, as indicated by the significantly higher rate of synonymous substitution (dS) over nonsynonymous substitutions (dN) across all sites (Table 1). However, there was no significant excess of synonymous substitutions in antigen-binding sites (ABS). Moreover, whereas dS did not differ significantly between ABS and non-ABS sites (Z = 0.355, p = 0.722), dN was considerably higher at ABS than at non-ABS sites (Z = 2.665, p = 0.008). This suggests that ABS possibly experienced positive selection but that the signal was obscured by purifying selection acting across sequences. However, the model of positive codon selection (M8) did not fit the data better (AIC = 1226.6) than the model of nearly neutral codon evolution (M7) (AIC = 1222.4, best).
We found significant linkage disequilibria between certain pairs of nucleotide alleles among putative loci (Table S2, Supplementary Materials), so it appeared prudent not to treat alleles from different clusters as independent predictors in a single analysis. Therefore, we performed separate analyses for clusters I and II, which were polymorphic in terms of amino acid sequences.
The effect of the MHC on the risk of infection
Because of the linkage disequilibria, alleles belonging to different clusters could not be treated as independent predictors. We therefore carried out separate analyses for two polymorphic clusters, II and III (see Fig. 1). Cluster I, which consisted of only one amino acid sequence that was present in all individuals, was excluded from the models.
We found a significant relationship between the presence of MHC alleles from both clusters and the prevalence of the blood parasite Babesia ssp. (Table 2). Allele AA*08 (cluster II) was negatively associated with the presence of the parasite: Babesia ssp. prevalence in animals carrying this allele was nearly half that of animals without it (13.9 and 25.7 %, respectively). The effect of allele AA*08 was, however, marginally not significant when we applied a correction for multiple comparisons (p = 0.063). In contrast, allele AA*05 (cluster III) had the opposite effect: the voles with this allele were infected more frequently than others (31.7 vs. 15.3 %). Because the alleles were not linked (Table S2, Suppl. Materials), we may assume that their effects independently predicted Babesia ssp. prevalence. Aside from genetic factors, Babesia ssp. infection was also associated with host body mass: infected animals weighed more than uninfected ones (34.8 g, SE = 0.60 vs. 27.3 g, SE = 1.85). There were no significant associations between presence of MHC alleles and infection with Mycoplasma ssp.
We found no effect of MHC alleles on infection by cestodes. The only factor retained in a minimal model of infections by the cestode Apostatandria was host body mass: the infected animals weighed more on average than uninfected ones (34.9 g, SE = 0.62 and 27.3 g, SE = 3.02 respectively) (χ2 = 8.236, p = 0.004). The minimal model of infection by the cestode Rodontolepis was the null model, and infection by nematodes was not analysed due to the low sample size.
The heterozygosity within clusters II and III was associated with the prevalence of Babesia ssp. (Table 3A). Animals heterozygous for cluster II were infected with Babesia ssp. less often than homozygotes (17.1 vs. 32.6 %), but at the same time, were more likely to have “resistance” allele AA*08 (7.3 vs. 40.01 %, G-test χ2 = 24.98, p < 0.001). Similarly, voles heterozygous for cluster III were infected more frequently (25 vs. 19 %) than homozygotes, and heterozygotes were also more likely to possess allele AA*05, which increased the risk of infection with Babesia ssp. (49.6 vs. 28.2. %, G test, χ2 = 5.962, p = 0.014). However, in a model accounting for the effects of alleles AA*05 and AA*08, the effect of heterozygosity at locus II remained significant, and heterozygosity at locus III became only marginally non-significant (Table 3B).
Effects of the prevalence of parasite infection and the MHC on survival
The low prevalence of nematodes did not allow for a determination of the effect of the Ivomec treatment on infection or the effect of nematodes on survival. Hence, the nematodes were not considered in subsequent analyses and we focused exclusively on blood parasites and cestodes.
Infection with Babesia ssp. significantly decreased winter survival (November to March, Table 4). Among infected animals, only 9.1 % survived, compared with 20.8 % survival of uninfected individuals. However, the effect of infection was more pronounced in the second part of winter: survival from January to March in the infected individuals was less than half that of uninfected ones (37 vs. 77 %, respectively). In contrast, none of the parasites affected survival in the first part of the winter (November to January, Table 4). Additionally, survival was significantly affected by the Ivomec treatment in the first half of the winter and overall (Table 4). The animals from the control group survived better than those injected with Ivomec (whole winter: 25 and 13 %, respectively; from November to January: 37.3 and 21.8 %, respectively). Similar trends were observed when we fitted the intensity of infection instead of parasite prevalence (Table S3, Suppl. materials). Neither the infection with cestodes, nor with Mycoplasma, affected the survival.
Winter survival was significantly affected by the presence of MHC DRB alleles from both clusters II and III (Table 5). The presence of allele AA*01 from cluster III increased the survival from November to March (27.5 % in carriers vs. 7.8 % in non-carriers) and from November to January (38.5 vs. 17.1 %). The allele also increased survival in the second part of the winter, from January to March (65 vs. 53 %), but the effect was not significant (Table 5). Among the alleles from cluster II, we found higher survival from November to March for the voles with allele AA*08; animals carrying the allele survived at approximately twice the rate of voles without the allele (44.9 vs. 22 %). The heterozygosity within clusters did not affect survival (Suppl. Materials, Table S4).
Discussion
Infections and survival
Parasites are believed to affect host population dynamics by increasing the mortality of infected individuals (Anderson and May 1979), which has been supported by numerous field studies (e.g. Gulland and Fox 1992; Burthe et al. 2008). However, parasites seldom represent the sole explanation for the increased mortality of infected animals (Telfer et al. 2010), and their effect on host populations often depends on other factors, such as host sex (Fuller and Blaustein 1996) or food availability (Murray et al. 1996). Here, we studied survival over the winter, which is the period when the tight energy budget of animals may make the negative effects of parasite infection particularly pronounced. We have demonstrated that infection with the blood parasite Babesia ssp., but not with cestodes, significantly decreases the winter survival of the root vole.
Blood parasites are generally considered harmful, as they destroy blood cells and may also damage other organs during their life cycle (Samuel et al. 2001). Blood parasites have been demonstrated to affect the survival of birds (Stjernman et al. 2008; de la Puente et al. 2010). Among mammals, decreased survival was recorded for malaria-infected polecats (Novilla et al. 1980) but not for a wild rodent, the degu, infected with Trypanosoma cruzi (Botto-Mahan et al. 2012). Babesia ssp. is a protozan, tick-borne parasite that attacks erythrocytes, which may lead to haemolytic anaemia (Irwin 2005). In voles, the infections are usually subclinical and persistent (Telfer et al. 2008). The risk of infection depends on coinfection with other parasite species (Telfer et al. 2010), however in the current paper, we found no such interactions.
The prevalence of nematodes in the studied root voles was surprisingly low compared with other reports on voles (e.g. Tollenaere et al. 2008; Janova et al. 2010; Kloch et al. 2010). Consequently, we could not assess the effect of gut nematodes on survival based on the effect of the anthelmintic treatment. However, we found that the animals injected with Ivomec survived at a lower rate than control individuals. Ivomec has been reported to be a safe drug for rodents, and we used the dose recommended in the literature (e.g., Wahid and Behnke 1996). However, our results indicated that treatment with Ivomec increased mortality. Recent findings have shown that the effect of anthelmintic treatment largely depends on the initial parasite burden; in wood mice, the survival of heavily infected individuals improved following the administration of an anthelmintic drug, but the effect on individuals with low nematode burdens was the opposite (Pedersen et al. 2011). Due to the low overall nematode prevalence, we hypothesise that a similar mechanism occurred in our population.
We found no effect of host body mass on survival, although heavier animals were infected with Babesia ssp. and Apostatandria more often. As the body mass of the root voles is strongly correlated with age (Borowski 2011), this effect may simply reflect the duration of exposure to infection; if the chance of encountering an infected vector or ingesting a cestode egg is constant, the risk of infection increases over time.
Effects of the MHC on infection and survival rates
The genes of the major histocompatibility complex are an important factor influencing host susceptibility to infections, and indeed, both negative and positive associations between the presence of particular MHC alleles and parasites have been shown in several vertebrate taxa, such as rodents (Meyer-Lucht and Sommer 2005; Froeschke and Sommer 2005; Kloch et al. 2010), ruminants (Paterson et al. 1998), fish (Eizaguirre et al. 2009) and birds (Westerdahl et al. 2005; Bonneaud et al. 2006; Loiseau et al. 2008). Similarly, in the present study, we detected a significant association between allele AA*05 and infection with Babesia ssp. Recently, Turner et al. (2011) showed that the susceptibility to Babesia microti in common vole was associated with the variation within genes coding interleukines Il2 and Il2b.
However, the fitness consequences of associations between parasites and immune genes are poorly understood. In the present paper, we have shown that infection with the blood parasite Babesia ssp. decreased winter survival in the root vole, but individuals with allele AA*08 tended to be infected less frequently and survived better than those without this allele. These associations are consistent with the existence of causal link between the MHC, parasites and survival in a wild rodent system. The presence of allele AA*01 also increased survival, particularly in the first part of the winter. However, we did not find an effect of the allele on any of the parasites. One possible explanation for this inconsistency is that the AA*01 allele may be associated with other pathogens not analysed in this study.
MHC genes form a large linkage group, so it remains uncertain whether the observed effects of DRB alleles on infection rates were due directly to this locus or to some other DRB-linked genes. We have not detected an unambiguous signal of historical selection on MHC DRB in the root vole. This locus appears to have evolved under purifying selection, and model selection procedures in PAML did not detect positive selection. Nevertheless, dN at ABS sites significantly exceeded dN at non-ABS sites, suggesting that positive selection did act on the codons involved in presenting the antigens.
Susceptibility to parasites may be affected not only by the effects of particular genes but also by individual MHC heterozygosity (Oliver et al. 2009), as heterozygotes are more likely to recognise parasite-derived antigens than homozygotes (Doherty and Zinkernagel 1975). MHC heterozygosity has been shown to be associated with decreased infection rates in both laboratory and field studies (McClelland et al. 2003; Froeschke and Sommer 2005), but several other studies have found no such association (Harf and Sommer 2005; Dionne et al. 2009). Ilmonen et al. (2007) found that wild-derived mice that were MHC heterozygotes did not show better resistance to Salmonella or better survival in semi-natural enclosures, even though earlier laboratory experiments documented a heterozygote advantage (Penn et al. 2002). These results emphasise the importance of studies on wild, outbred species.
In the present study, depending on the locus, we found either an increased or a decreased probability of infection of heterozygotes with Babesia ssp. Closer examination revealed that this apparent inconsistency can be partly explained by heterozygous animals being more likely to possess either “resistance” or “susceptibility” alleles. Heterozygosity in cluster III was associated with a decreased risk of infection with the blood parasite Babesia ssp. because this cluster included allele AA*08, which was present more often in uninfected animals. Similarly, animals that were heterozygous in cluster II were infected more frequently, as they were more likely to possess the “susceptible” allele AA*05. Nevertheless, the contrasting effects of heterozygosity between the two clusters remained marginally significant after the effects of alleles AA*05 and AA*08 were accounted for in a statistical model (Table 3B), possibly due to the cumulative effects of the other alleles, which were too weak to become individually significant.
Conclusions
In the present paper, we found significant effects of the blood parasite Babesia ssp. on survival. The prevalence of the parasite as well as the survival rate were correlated with the presence of MHC alleles, providing strong evidence for contemporary, parasite-driven selection acting on MHC genes. Overall, our results demonstrate that infection with Babesia ssp. is a strong predictor of winter survival and that MHC genes are important predictors of infection status as well as survival in the root vole.
References
Anderson RM, May RM (1979) Population biology of infectious disease: part I. Nature 280:361–367
Babik W, Taberlet P, Ejsmond MJ, Radwan J (2009) New generation sequencers as a tool for genotyping of highly polymorphic multilocus MHC system. Mol Ecol Res 9:713–719
Bajer A, Pawełczyk A, Behnke JM, Gilbert FS, Siński E (2001) Factors affecting the component community structure of haemoparasites in bank voles (Clethrionomys glareolus) from the Mazury Lake District region of Poland. Parasitology 1:43–54
Banks SC, Dubach J, Viggers KL, Lindenmayer DB (2010) Adult survival and microsatellite diversity in possums: effects of major histocompatibility complex-linked microsatellite diversity but not multilocus inbreeding estimators. Oecologia 162:359–370
Benjamini Y, Yekutieli D (2001) The control of the false discovery rate in multiple testing under dependency. Ann Statist 29:1165–1188
Bernatchez L, Landry C (2003) MHC studies in nonmodel vertebrates: what have we learned about natural selection in 15 years. J Evol Biol 16:363–377
Bodmer W (1972) Evolutionary significance of the HL-A system. Nature 237:139–145
Bonneaud C, Perez-Tris J, Federici P, Chastel O, Sorci G (2006) Major histocompatibility alleles associated with local resistance to malaria in a passerine. Evolution 60:383–389
Borghans JAM, Beltman JB, De Boer RJ (2004) MHC polymorphism under host–pathogen coevolution. Immunogenetics 55:732–739
Borkowska A, Borowski Z, Krysiuk K (2009) Multiple paternity in free-living root voles (Microtus oeconomus). Behav Process 82:211–213
Borowski Z (2002) Individual and seasonal differences in antipredatory behaviour of root voles: a field experiment. Can J Zool 80:1520–1525
Borowski Z (2011) The impact of predation on small rodent population dynamics as exemplified by the root vole (Mictotus oeconomus). Papers of the Forest Research Institute. ISBN 978-83-87647-98-8
Botto-Mahan C, Bacigalupo A, Correa JP, Oda E, Solari A (2012) Field assessment of Trypanosoma cruzi infection and host survival in the native rodent Octodon degus. Acta Trop 122:164–167
Brouwer L, Barr I, van de Pol M, Burke T, Komdeur J, Richardson DS (2010) MHC-dependent survival in a wild population: evidence for hidden genetic benefits gained through extra-pair fertilizations. Mol Ecol 19:3444–3455
Burthe S, Telfer S, Begon M, Bennett M, Smith A, Lambin X (2008) Cowpox virus infection in natural field vole Microtus agrestis populations: significant negative impacts on survival. J Anim Ecol 77:110–119
Crawley MJ (2007) The R book. Wiley, Chichester
de la Puente JM, Merino S, Tomás G, Moreno J, Morales J, Lobato E, García-Fraile S, Belda EJ (2010) The blood parasite Haemoproteus reduces survival in a wild bird: a medication experiment. Biol Lett 6:63–665
Dionne M, Miller KM, Dodson JJ, Bernatchez L (2009) MHC standing genetic variation and pathogen resistance in wild Atlantic salmon. Phil Trans R Soc B 364:1555–1565
Doherty PC, Zinkernagel RM (1975) Enhanced immunological surveillance in mice heterozygous at H-2 gene complex. Nature 256:50–52
Dunn A, Keymer A (1986) Factors affecting the reliability of the McMaster technique. J Helminthol 60:260–262
Eizaguirre C, Yeates SE, Lenz TL, Kalbe M, Milinski M (2009) MHC-based mate choice combines good genes and maintenance of MHC polymorphism. Mol Ecol 18:3316–3329
Froeschke G, Sommer S (2005) MHC class II DRB variability and parasite load in the striped mouse (Rhabdomys pumilio) in the southern Kalahari. Mol Biol Evol 22:1254–1259
Fuller CA, Blaustein R (1996) The effect of the intestinal parasite Eimeria arizonensis on survival of deer mice (Peromyscus maniculatus) in natural and semi-natural populations. Ecology 77:2196–2202
Galan M, Guivier E, Caraux G, Charbonnel N, Cosson JF (2010) A 454 multiplex sequencing method for rapid and reliable genotyping of highly polymorphic genes in large-scale studies. BMC Genomics 11:296
Gannon WL, Sikes RS, The Animal Care and Use Comittee of ASM (2007) Guidelines of the American Society of Mammologists for the use of wild mammals in research. J Mammol 88:809–823
Garnier-Gere P, Dillmann C (1992) A computer program for testing pairwise linkage disequilibria in subdivided populations. J Heredity 83:239
Gulland FMD, Fox M (1992) Epidemiology of nematode infection of Soay sheep (Ovis aries L.) on St. Kilda. Parasitology 105:481–492
Harf R, Sommer S (2005) Association between major histocompatibility complex class II DRB alleles and parasite load in the hairy–footed gerbil, Gerbillurus paeba, in the southern Kalahari. Mol Ecol 14:85–91
Hedrick PW (2002) Pathogen resistance and genetic variation at MHC loci. Evolution 56:1902–1908
Hedrick PW, Kim TJ, Parker KM (2001) Parasite resistance and genetic variation in the endangered Gila topminnow. Anim Conserv 4:103–109
Hill AVS, Allsopp CEM, Kwiatkowski D, Anstey NM, Twumasi P, Rowe PA, Bennett S, Brewster D, McMichael AJ, Greenwood BM (1991) Common West African HLA antigens are associated with protection from severe malaria. Nature 352:595–600
Hughes AL, Nei M (1988) Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature 335:167–170
Ilmonen P, Penn DJ, Damjanovich K, Morrison L, Ghotbi L, Potts WK (2007) Major histocompatibility complex heterozygosity reduces fitness in experimentally infected mice. Genetics 176:2501–2508
Irwin P (2005) Babesiosis and cytauxzoonosis. In: Shaw SE, Day MJ (eds) Arthropod-borne infectious diseases of the dog and cat. Manson Publishing, London, pp 63–77
Janova E, Skoric M, Heroldova M, Tenora F, Fictum P, Pavlik I (2010) Determinants of the prevalence of Heligmosomum costellatum (Heligmosomidae: trichostrongyloidea) in a common vole population in southern Moravia Czech Republic. J Helminthol 84:410–414
Jędrzejewska B,Pucek Z, Jędrzejewski W (2004) Seed crops and forest rodents. In: Jędrzejewska B, Wójcik JM (eds) Essays on mammals of białowieża forest, Mammal Research Institute Białowieża, pp 129–139
Kloch A, Babik W, Bajer A, Sinski E, Radwan J (2010) Effects of an MHC-DRB genotype and allele number on the load of gut parasites in the bank vole Myodes glareolus. Mol Ecol 19:255–265
Loiseau C, Zoorob R, Garnier S, Birard J, Federici P, Julliard R, Sorci G (2008) Antagonistic effects of a MHC class I allele on malaria–infected house sparrows. Ecol Lett 11:258–265
Loiseau C, Zoorob R, Robert A et al (2011) Plasmodium relictum infection and MHC diversity in the house sparrow (Passer domesticus). Proc Roy Soc, Ser B-Biol Sci 278:1264–1272
Marsh SGE, Parham P, Barber LD (2000) The HLA facts book. Academic Press, London
McClelland EE, Penn DJ, Potts WK (2003) Major histocompatibility complex heterozygote superiority during coinfection. Infect Imm 71:2079–2086
Meyer-Lucht Y, Sommer S (2005) MHC diversity and the association to nematode parasitism in the yellow–necked mouse (Apodemus flavicollis). Mol Ecol 14:2233–2243
Murray DL, Keith LB, Cary JR (1996) The effect of anthelmintic treatment on the parasite burden of snowshoe hares. Can J Zool 74:1604–1611
Narum S (2006) Beyond Bonferroni: less conservative analyses for conservation genetics. Cons Genet 7:783–787
Norrdahl K, Korpimäki E (2005) Survival through bottlenecks of vole cycles: refuge or chance events? Evol Ecol 19:339–361
Novilla MA, Carpenter JW, Kwapein RP (1980) Dual infection of Siberian polecats with Encephalitozoon cuniculi and Hepatozoon mustelis sp. In: Montali RJ (ed) Symposium on the comparative pathology of zoo animals National Zoological Park. Smithsonian Institution Press, Washington, pp 353–363
Oliver MK, Telfer S, Piertney SB (2009) Major histocompatibility complex (MHC) heterozygote superiority to natural multi-parasite infections in the water vole (Arvicola terrestris). Proc Biol Sci 276:1119–1128
Paterson S, Wilson K, Pemberton JM (1998) Major histocompatibility complex variation associated with juvenile survival and parasite resistance in a large unmanaged ungulate population. Proc Natl Acad Sci USA 95:3714–3719
Pedersen AB, Knowles SCL, Petchey OL, Fenton A (2011) The role of parasite community structure in mediating the success of anthelmintic treatment strategies. Annual spring meeting British society for parasitology, Nottingham, April 2011
Penn DJ, Damjanovich K, Potts WK (2002) MHC heterozygosity confers a selective advantage against multiple-strain infections. Proc Natl Acad Sci USA 99:11260–11264
Piertney SB, Oliver MK (2006) The evolutionary ecology of the major histocompatibility complex. Heredity 96:7–21
Posada D, Buckley TR (2004) Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst Biol 53:793–808
Potts WK, Wakeland EK (1990) The maintenance of MHC polymorphism. Immunol Today 11:39–40
R Development Core Team (2008) R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing, Vienna Austria, http://www.r-project.org
Reche PA, Reinherz EL (2003) Sequence variability analysis of human class I and class II MHC molecules: functional and structural correlates of amino acid polymorphisms. J Mol Biol 331:623–641
Robinson J, Waller MJ, Parham P, de Groot N, Bontrop R, Kennedy LJ, Stoehr P, Marsh SGE (2003) IMGT/HLA and IMGT/MHC: sequence databases for the study of the major histocompatibility complex. Nucl Acid Res 31:311–314
Rousset F (2008) Genepop v007: a complete reimplementation of the Genepop software for Windows and Linux. Mol Ecol Res 8:03–106
Rozera G, Abbate I, Bruselles A, Vlassi C, D’Offizi G, Narciso P, Chillemi G, Prosperi M, Ippolito G, Capobianchi MR (2009) Massively parallel pyrosequencing highlights minority variants in the HIV-1 env quasispecies deriving from lymphomonocyte sub-populations. Retrovirology 12:6–15
Samuel WM, Pybus MJ, Kocan AA (eds) (2001) Parasitic diseases of wild mammals, 2nd edn. Iowa State Press, Ames
Savage AE, Zamudio KR (2011) MHC genotypes associate with resistance to a frog-killing fungus. Proc Natl Acad Sci USA 108:16705–16710
Scantlebury M, Waterman JM, Hillegass MA, Speakman JR, Bennett NC (2007) Energetic costs of parasitism in the Cape ground squirrel Xerus inauris. Proc R Soc B 274:2169–2177
Schaschl H, Suchentrunk F, Morris DL, Ben Slimen H, Smith S, Arnold W (2012) Sex-specific selection for MHC variability in Alpine chamois. BMC Evol Biol 12:20
Smulders MJM, Snoek LB, Booy G, Vosman B (2003) Complete loss of MHC genetic diversity in the common hamster (Cricetus cricetus) population in the Netherlands Consequences for conservation strategies. Conserv Genet 4:441–451
Sommer S (2005) The importance of immune gene variability (MHC) in evolutionary ecology and conservation. Front Zool 2:16
Spurgin LG, Richardson DS (2010) How pathogens drive genetic diversity: MHC mechanisms and misunderstandings. Proc R Soc B 277:979–988
Stjernman M, Råberg L, Nilsson J-Å (2008) Maximum host survival at intermediate parasite infection intensities. PLoS ONE 3(6):e2463. doi:101371/journalpone0002463
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Telfer S, Birtle R, Bennett M, Lambin X, Paterson S, Begon M (2008) Parasite interactions in natural populations: insights from longitudinal data. Parasitology 135:767–781
Telfer S, Lambin X, Birtles R, Beldomenico P, Burthe S, Paterson M, Begon M (2010) Species interactions in a parasite community drive infection risk in a wildlife population. Science 330:243–246
Thienpont D, Rochette F, Vanparijs OFJ (1986) Diagnosing helminthiasis by coprological examination. Janssen Research Foundation, Beerse
Tollenaere C, Bryja J, Galan M, Cadet P, Deter J, Chaval Y, Berthier K, Ribas SA, Voutilainen L, Laakkonen J, Henttonen H, Cosson JF, Charbonnel N (2008) Multiple parasites mediate balancing selection at two MHC class II genes in the fossorial water vole: insights from multivariate analyses and population genetics. J Evol Biol 21:1307–1320
Trowsdale J (1995) Both man and bird and beast–comparative organization of MHC genes. Immunogenetics 41:1–17
Turner AK, Begon M, Jackson JA, Bradley JE, Paterson S (2011) Genetic diversity in cytokines associated with immune variation and resistance to multiple pathogens in a natural rodent population. PLoS Genet 7:10
Wahid FN, Behnke JM (1996) Genetic control of acquired resistance to Heligmosomoides polygyrus: overcoming genetically determined weak responder status by strategic immunization with ivermectin-abbreviated infections. J Helminthol 70:159–168
Wedekind C, Walker M, Portmann J, Cenni B, Muller R, Binza T (2004) MHC-linked susceptibility to a bacterial infection but no MHC-linked cryptic female choice in whitefish. JEvol Biol 17:11–18
Westerdahl H, Waldenström J, Hansson B, Hasselquist D, von Schantz T, Bensch S (2005) Associations between malaria and MHC genes in a migratory songbird. Proc Biol Sci 272:1511–1518
Worley K, Collet J, Spurgin LG, Cornwallis C, Pizzari T, Richardson DS (2010) MHC heterozygosity and survival in red junglefowl. Mol Ecol 19:3064–3075
Yang Z (1997) PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci 13:555–556
Zagalska-Neubauer M, Babik W, Stuglik M, Gustafsson L, Cichoń M, Radwan J (2010) 454 sequencing reveals extreme complexity of the class II Major Histocompatibility Complex in the collared flycatcher. BMC Evol Biol 10:395
Acknowledgments
The authors would like to thank Stuart Piertney and anonymous reviewers for comments on previous versions of the manuscript, prof. Jan Taylor who kindly provided us access to the field station in Gugny of the University of Białystok, and to the Functional Genomics Center, Uni/ETH Zurich, especially to Marzanna Kuenzli, for running our amplicons on 454. We thank the following persons for data collection in the field: D. Bugajna, D. Dudek, Z. Jędrzejewski-Szmek, B. Jurgiel, Ł. Korczewski, J. Leś, A. Makal, O. Michalski, Z. Prokop, M. Sobczyk, P. Szafrańska. The work was supported by Polish Ministry of Science grant no. N N304 168836, and by Jagiellonian University funds DS/WBINOZ/INOŚ/762/10.
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License (https://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Kloch, A., Baran, K., Buczek, M. et al. MHC influences infection with parasites and winter survival in the root vole Microtus oeconomus . Evol Ecol 27, 635–653 (2013). https://doi.org/10.1007/s10682-012-9611-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10682-012-9611-1