Abstract
The aim of this study was to assess genotype by environment interaction for seed quality traits in interspecific cross-derived Brassica lines by the additive main effects and multiplicative interaction (AMMI) model. The study comprised of 25 winter rapeseed genotypes i.e.: B. napus cultivar Californium, twenty three cross-derived Brassica lines and male sterile line of an F8 generation of B. napus (MS8), selected from resynthesized oilseed rape (B. rapa ssp. chinensis × B. oleracea var. gemmifera) using in vitro cultures of isolated embryos. Field trials were performed at three locations in 3 years in a randomized complete block design, with three replicates. AMMI analyses revealed significant genotype and environmental effects as well as genotype by environmental interaction with respect to all five observed traits. The lines 16 (B. napus line MS8 × B. rapa ssp. pekinensis) and 7 (B. napus line MS8 × B. carinata) are recommended for further inclusion in the breeding programs because their stability and good average values of observed traits, except total glucosinolates content for line 16 (the best total genotype selection indexes were equal to 81 and 97, respectively).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Brassica L. is a diverse genus that includes six species cultivated as important crops, which yield edible oil and condiment seeds, roots, leaves, stems, vegetative and floral buds and meristems. Due to their wide adaptation and ability to thrive under varying agroclimatic conditions, brassica crops are grown throughout the world for food, animal forage, fodder and industrial applications (Eckes et al. 2017). In Poland within Brassica species, winter oilseed rape is the most important oil–protein plant (Liersch et al. 2016; Szała et al. 2016; Sosnowska et al. 2017).
Given the importance of Brassica species worldwide, integrated approaches to research and plant breeding for crop improvement are required to address future global challenges, such as satisfying increased demand for higher quality and nutritious food, the reduction of waste, and producing predictable yields in a more variable environment. Researchers have evaluated many aspects of Brassica phenotypic diversity to assist in crop improvement, including yield, resistance to pathogens and pests and the composition of industrially important compounds.
Nowadays, development of oilseed rape cultivars with good quality i.e. high oil, protein as well as low erucid acid, glucosinolate and fiber content is a challenging objective for many rapeseed breeding programs (Bocianowski et al. 2014; Niemann et al. 2012; Zou et al. 2016). It is known that all these seed quality traits are influenced by genetic and environmental factors, so their precise estimation requires phenotyping in replicated multi-environmental field trials.
In field crop trials, genotype-by-environment (GE) interaction is often analyzed by the additive main effects and multiplicative interaction (AMMI) model (Zobel et al. 1988). The AMMI model is a combines the analysis of variance (ANOVA) with additive parameters and the principal component analysis (PCA) with multiplicative parameters in a single analysis. The AMMI biplot graphic display simultaneously both main and interaction effects for genotypes and environments, and enables a single analysis of the genotype by environment interaction. The AMMI is, therefore, also known as interaction PCA (Gauch and Zobel 1990).
The aim of this paper was to assess genotype by environment interaction for five quantitative traits (oil content, protein content, total glucosinolates content, acid detergent fiber and neutral detergent fiber) in interspecific cross-derived Brassica lines grown in three locations in the Greater Poland by the AMMI model.
Materials and methods
Plant material for field trials consisted of 25 winter rapeseed genotypes i.e.: B. napus cv. Californium, twenty three interspecific cross-derived Brassica lines and male sterile line of an F8 generation of B. napus (MS8). MS8 line was selected from resynthesized oilseed rape (B. rapa ssp. chinensis × B. oleracea var. gemmifera) using in vitro cultures of isolated embryos (Table 1). F5–F7 generation of tested lines as well as control genotypes i.e. B. napus cv. Californium and MS8 line were selected from the rapeseed breeding program of the Department of Genetics and Plant Breeding, Poznań University of Life Sciences (PULS). The study was carried out in three locations i.e. the PULS experimental station Dłoń (51°41′23″N, 17°04′10″E) located 100 km south of Poznań, Złotniki (52°25′18″N, 16°50′41″E) located 15 km west from Poznań and in the experimental fields in Poznań–Sołacz (52°25′18″N, 16°54′26″E). The field trials at all locations were arranged in a randomized complete block design with three replicates. Each genotype was grown in a three row plot of 9.0 m2 with a 0.30 row distance and a sawing density of 60 seeds/m2. Agricultural practices were optimal for local agroecological conditions in all investigated locations. Plots were harvested using a plot harvester. These locations have different types of soil and weather conditions. The field experiment in Dłoń was conducted on typical heavy soil of III quality class. The field experiment in Sołacz was conducted on typical light, sandy soil of IV quality class. The field experiment in Złotniki was conducted on typical luvisols soil of IVa quality class. Complex of agricultural usefulness were: good rye in Dłoń, good wheat in Sołacz and very good rye in Złotniki. In crop seasons 2010/2011, 2011/2012 and 2012/2013 weather conditions were normal for Poland. The sums of precipitation during vegetation season of winter oilseed rape in 2011, 2012 and 2013 were, respectively, in Dłoń: 465, 631 and 565 mm, in Sołacz: 477, 514 and 607 mm, in Złotniki: 402, 678 and 581 mm. Mean annual temperature during vegetation season of winter oilseed rape in 2011, 2012 and 2013 were, respectively, in Dłoń: 10.0, 10.1 and 9.7 °C, in Sołacz: 9.6, 9.4 and 9.2 °C, in Złotniki: 15.2, 9.1 and 9.8 °C.
Quality analysis
The seed samples for the analysis of protein content (in %), oil content (in %), total glucosinolates content (in umol/gsm) and fiber content (in %) were collected from five self-pollinated plants from each tested line in each replication. The fiber content in the cross-derived lines and control seeds was expressed as the average value of the neutral detergent fiber (NDF) and the acid detergent fiber (ADF) as a subfraction of the NDF. The seed quality traits content was determined by near-infrared reflectance spectroscopy (NIRS) (Velasco et al. 1999). These analyses were performed at the Małyszyn Laboratory that belongs to Plant Breeding Strzelce in triplicate. For NIRS analysis minimum 3 g of intact seeds were used. Every seed probe was scanned on the NIRS Systems monochromator model 6500 (NIR Systems, Inc. Silver Springers, MD, USA).
Statistical analysis
A two-way fixed effect model was fitted to determine the magnitude of the main effects of variation and their interaction on each trait. Least-squares means were simultaneously produced for the AMMI model. The model first fits additive effects for the main effects of genotypes (G) and environments (E) followed by multiplicative effects for GE interaction by principal component analysis. Environments were defined as combinations of locations and years (E1—Dłoń, 2011; E2—Poznań–Sołacz, 2011; E3—Złotniki, 2011; E4—Dłoń, 2012; E5—Poznań–Sołacz, 2012; E6—Złotniki, 2012; E7—Dłoń, 2013; E8—Poznań–Sołacz, 2013; E9—Złotniki, 2013). The AMMI model (Gauch and Zobel 1990; Nowosad et al. 2016) is given by:
where yge is the trait mean of genotype g in environment e, μ is the grand mean, αg is the genotypic mean deviations, βe is the environmental mean deviations, N is the number of PCA axis retained in the adjusted model, λn is the eigenvalue of the PCA axis n, γgn is the genotype score for PCA axis n, δen is the score eigenvector for PCA axis n, Qge is the residual, including AMMI noise and pooled experimental error. Expected distribution of Qge is normal. The AMMI stability values (ASVs) were used to compare the stability of genotypes as described by Purchase et al. (2000):
where SS was the sum of squares, IPCA1 and IPCA2—the first and the second interaction principal component axes, respectively; the IPCA1 and IPCA2 scores were the genotypic scores in the AMMI model. ASV was the distance from zero in a two-dimensional scatterplot of IPCA1 scores against IPCA2 scores. Since the IPCA1 score contributes more to GE sum of square, it has to be weighted by the proportional difference between IPCA1 and IPCA2 scores to compensate for the relative contribution of IPCA1 and IPCA2 total GE sum of squares. The higher the IPCA score, either negative or positive, the more specifically adopted a genotype is to certain environments. Lower ASV score indicate a more stable genotype across environments (Nowosad et al. 2017).
Genotype selection index (GSI) was calculated for each genotype which incorporates both mean of trait and ASV index in single criteria (GSIi) as (Farshadfar and Sutka 2003)
where GSIi is genotype selection index for ith genotype, RMi is rank of trait mean (from maximal to minimal for oil content and protein content, and from minimal to maximal for total glucosinolates content, ADF and NDF) for ith genotype, RAi is rank of the AMMI stability value for the ith genotype. Finally, total genotype selection index (TGSI) was calculated for each genotype as sum of GSIs for all five traits of study. All the analyses were conducted using the GenStat v. 18 statistical software package.
Results
Oil content
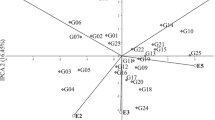
The three sources of variation were highly significant. In the analysis of variance, the sum of squares for environment main effect represented 16.69% of the total oil content variation. The differences between genotypes explained 9.37% of the total oil content variation, while the effects of GE interaction explained 25.70% (Table 2). Values for the three principal components were also highly significant and accounted jointly for 85.38% of the whole effect it had on the variation of oil content. The first principal component (IPCA 1) accounted for 51.68% of the variation caused by interaction, while IPCA 2 and IPCA 3 accounted for 18.23 and 15.47%, respectively. Genotype stability is considered as non significant reaction to changing environmental conditions, agronomic factors, weather conditions, biotic and abiotic stresses. In this study, climatic conditions were the source of this variation component. The stability of tested genotypes can be evaluated according to biplot for oil content (Fig. 1). Interspecific Brassica cross-derived lines interacted differently with climate conditions in the observed environments. The lines 2, 4, 5, 12 and 13 interacted positively with the E2, E5, E6 and E8 environments, but negatively with the E4 (Fig. 1). The lines 20, 21, 22, 23, 24 and 25 interacted positively with the E1 and E7 environments, but negatively with the E3 and E9. The analysis showed that some genotypes have high adaptation; however, most of them have specific adaptability. AMMI stability values (ASV) revealed variations in oil content stability among the 25 genotypes (Table 3). According to Purchase et al. (2000), a stable variety is defined as one with ASV value close to zero. Consequently, the lines 10 and 1 with ASV of, respectively, 0.271 and 0.476, were the most stable, while the lines 9 and 3 were the least stable (Table 3). The line 16 with the highest average oil content (44.20%) and ASV equal to 0.939 is line with the best genotype selection index (7). A group of lines: 7, 8, 11 and 19 had high averages of oil content and show high stability (Fig. 1, Table 3). These lines had good genotype selection index, respectively, 11, 11, 11 and 12 (Table 3).
Biplot for genotype by environment interaction of oil content in interspecific cross-derived Brassica lines in nine environments, showing the effects of primary and secondary components (IPCA 1 and IPCA 2, respectively) (E1—Dłoń, 2011; E2—Poznań–Sołacz, 2011; E3—Złotniki, 2011; E4—Dłoń, 2012; E5—Poznań–Sołacz, 2012; E6—Złotniki, 2012; E7—Dłoń, 2013; E8—Poznań–Sołacz, 2013; E9—Złotniki, 2013)
Protein content
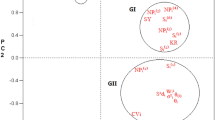
The three sources of variation for the protein content were highly significant. In the ANOVA, the sum of squares for environment main effect represented 50.08% of the total protein content variation. The differences between genotypes explained 10.03% of the total oil content variation, while the effects of GE interaction explained 23.05% (Table 2). Values for the three principal components were also highly significant and accounted jointly for 86.40% of the whole effect it had on the variation of protein content. The first principal component (IPCA 1) accounted for 54.68% of the variation caused by interaction, while IPCA 2 and IPCA 3 accounted for 19.01 and 12.71%, respectively. The stability of tested genotypes can be evaluated according to biplot for protein content (Fig. 2). The lines 2, 3, 5, 6, 18 and 19 interacted positively with the E1 and E4 environments, but negatively with the E3 and E6 (Fig. 2). The lines 9 and 14 interacted positively with the E7 environment, but negatively with the E9. The line 1 interacted positively with the E2, E5 and E8 environments. The lines 6, 1 and 17 with ASV of, respectively, 0.352, 0.410 and 0.454, were the most stable, while the lines 3 and 9 were the least stable (Table 3). The lines 6 and 16 with high average protein content (21.34% and 21.37%, respectively) and ASV equal to 0.352 and 0.454, respectively, are lines with the best genotype selection index (6).
Biplot for genotype by environment interaction of protein content in interspecific cross-derived Brassica lines in nine environments, showing the effects of primary and secondary components (IPCA 1 and IPCA 2, respectively) (E1—Dłoń, 2011; E2—Poznań–Sołacz, 2011; E3—Złotniki, 2011; E4—Dłoń, 2012; E5—Poznań–Sołacz, 2012; E6—Złotniki, 2012; E7—Dłoń, 2013; E8—Poznań–Sołacz, 2013; E9—Złotniki, 2013)
Total glucosinolates content
In the ANOVA, the sum of squares for genotypes main effect represented 77.63% of the total, and this factor had the highest effect on the total glucosinolates content. The differences between environments explained 1.07% of the total glucosinolates content variation, while the effects of GE interaction explained 3.78% (Table 2). Values for the three principal components were also highly significant. The three principal components of GE interaction accounted jointly for 82.82% of the whole effect it had on the variation of total glucosinolates content. The first principal component (IPCA 1) accounted for 54.89% of the variation caused by interaction, IPCA 2 accounted for 14.91% while IPCA 3 accounted for 13.02% (Fig. 3, Table 2). The stability of tested lines can be evaluated according to biplot for total glucosinolates content (Fig. 3). The lines 6, 19, 20 and 21 interacted positively with the E4, but negatively with the E3, E6 and E9 environments (Fig. 3). The lines 15, 22, 23 and 24 interacted positively with the E7, but negatively with the E3. The lines 14, 10 and 18 with ASV of, respectively, 0.346, 0.517 and 0.546 were the most stable, while the lines such as 6, 17, 21 and 24 were the smallest stable (Table 3). A group of lines: 11, 13, 10 and 1 had the least averages of total glucosinolates content. The line 10 has the best genotype selection index, equal to 5 (Table 3). However, the line 6 has the worst genotype selection index (50).
Biplot for genotype by environment interaction of total glucosinolates content in interspecific cross-derived Brassica lines in nine environments, showing the effects of primary and secondary components (IPCA 1 and IPCA 2, respectively) (E1—Dłoń, 2011; E2—Poznań–Sołacz, 2011; E3—Złotniki, 2011; E4—Dłoń, 2012; E5—Poznań–Sołacz, 2012; E6—Złotniki, 2012; E7—Dłoń, 2013; E8—Poznań–Sołacz, 2013; E9—Złotniki, 2013)
ADF
The three sources of variation were highly significant. In the ANOVA, the sum of squares for environment main effect represented 41.98% of the total, and this factor had the highest effect on ADF (Table 2). The differences between genotypes explained 12.95% of the total ADF variation, while the effects of GE interaction explained 29.62%. Values for the three principal components were also highly significant. The three principal components of GE interaction accounted jointly for 93.14% of the whole effect it had on the variation of ADF. The first principal component (IPCA 1) accounted for 50.22% of the variation caused by interaction, while IPCA 2 and IPCA 3 accounted for 34.74 and 8.18%, respectively. The lines 3 and 9 interacted positively with the E3 and E9 environments, but negatively with the E1 and E7 (Fig. 4). The lines 2, 5 and 12 interacted positively with the E6 environment, but negatively with the E4. The highest stability on ADF we observed for line 19 (ASV = 0.105). The best genotype selection index, equal to 9, has line 13, with small ADF (20.46) and ASV = 0.236 (Table 3).
Biplot for genotype by environment interaction of ADF in interspecific cross-derived Brassica lines in nine environments, showing the effects of primary and secondary components (IPCA 1 and IPCA 2, respectively) (E1—Dłoń, 2011; E2—Poznań–Sołacz, 2011; E3—Złotniki, 2011; E4—Dłoń, 2012; E5—Poznań–Sołacz, 2012; E6—Złotniki, 2012; E7—Dłoń, 2013; E8—Poznań–Sołacz, 2013; E9—Złotniki, 2013)
NDF
The sum of squares for environment main effect represented 22.92% of the total NDF variation. The differences between genotypes explained 33.39% of the total NDF variation, while the effects of GE interaction explained 21.41% (Table 2). Values for the three principal components were also highly significant and accounted jointly for 89.40% of the whole effect it had on the variation of NDF. The IPCA 1 accounted for 38.86% of the variation caused by interaction, while IPCA 2 and IPCA 3 accounted for 26.87 and 23.67%, respectively. The lines 4, 6 and 14 interacted positively with the E3, E6 and E9 environments, but negatively with the E1 and E7 (Fig. 5). The lines 17, 22, 24 and 25 interacted positively with the E1 and E7 environments, but negatively with the E3, E4, E6 and E9. The lines 23, 19 and 10 with ASV of, respectively, 0.377, 0.392 and 0.394, were the most stable, while the lines 6 and 3 were the least stable (Table 3). The line 16 with small average NDF (24.10%) and ASV equal to 0.506 is line with the best genotype selection index (9).
Biplot for genotype by environment interaction of NDF in interspecific cross-derived Brassica lines in nine environments, showing the effects of primary and secondary components (IPCA 1 and IPCA 2, respectively) (E1—Dłoń, 2011; E2—Poznań–Sołacz, 2011; E3—Złotniki, 2011; E4—Dłoń, 2012; E5—Poznań–Sołacz, 2012; E6—Złotniki, 2012; E7—Dłoń, 2013; E8—Poznań–Sołacz, 2013; E9—Złotniki, 2013)
The best total genotype selection index we observed for line 16 (TGSI = 81) and line 7 (TGSI = 97), while the worst—line 9 (TGSI = 187). The lines 16 (B. napus line MS8 × B. rapa ssp. pekinensis) and 7 (B. napus line MS8 × B. carinata) are recommended for further inclusion in the breeding programs because of their stability and good average values of observed traits, except total glucosinolates content for line 16.
Discussion
In Brassica species and especially in Brassica napus breeding, traits related to commercial success are of highest importance (Friedt and Snowdon 2010). However, such traits in many cases can only be assessed in an advanced developmental stage (Körber et al. 2016). The oil content, protein content, total glucosinolates content, ADF and NDF in winter rapeseed are traits determined by multiple genes that cause change in the performance of genotypes depending on the cultivation environment. That is why the importance of GE interactions in plant breeding programs have been a focus of attention for breeders (Golkari et al. 2016; Nowosad et al. 2016; Chen et al. 2017). Genotype by environment interaction is an important and challenging issue for plant breeders especially in developing new improved varieties. Multi-environment trials are used to determine sites representing the target environment and can identify superior lines or cultivars for recommendation to breeders (Golkari et al. 2016). Data collected from such trials are needed for precise estimation of genotype value and yield stability (Yan and Hunt 2001). These trials facilitate quantification of the environment and GE interactions. Differences in environmental conditions may cause large GE interactions, especially under drought-prone environments. The interpretation of GE interactions can be facilitated using several statistical models.
Traditional, statistical methods as linear regression ANOVA and PCA are often not effective for understanding and evaluating complex data from multi-environments yield trails. In contrast to the standard statistical analyses, AMMI incorporates ANOVA and PCA into a single model and enables simple visual interpretation of the GE interaction. AMMI model is usually constructed from the first two IPCA axes. Measuring GE interaction is very important to determine an optimum breeding strategy for releasing genotypes with an adequate adaptation to target environments (Fox et al. 1997). The AMMI model was often used in study of many species (Abakemal et al. 2016; Edwards 2016; Nowosad et al. 2016). The AMMI model provides a useful tool in diagnosing GE interaction patterns and improving the accuracy of response estimates. It enables clustering of genotypes based on similarity of response characteristics and identifying potential trends across environments (Bocianowski et al. 2018). The suggested strategy could extract more information from the GE interaction, thereby aiding researchers in identifying specific genotypes with competitive yields across diverse environments. The genotype and environment main effects as well as GE interaction had the strongest effect on all five observed traits (oil content, protein content, total glucosinolates content, ADF, NDF) expression in Greater Poland. AMMI analyses permits estimation of interaction effect of a genotype in each environment and it helps to identify genotypes best suited for specific environmental conditions. AMMI analyses revealed significant GE interaction with respect to all five traits. The AMMI stability value exposed high genotypes stability. The AMMI results displayed on the GE biplot enables determination of the main effect of the genotype, the environment, and the most meaningful GE interactions.
In general, environments with scores near zero have little interaction across genotypes and provide low discrimination among genotypes (Anandan et al. 2009). In this study, these patterns were observed in E2, E5 and E8 environments (Figs. 1, 2, 3, 4, 5). AMMI models are capable of measuring the weight of the environments, the genotypes and their interactions throughout a value that measures how stable a genotype is in all environments in terms of particular observed traits. The lines 16 (B. napus line MS8 × B. rapa ssp. pekinensis) and 7 (B. napus line MS8 × B. carinata) are recommended for further inclusion in the breeding programs because their stability and good average values of observed traits, except total glucosinolates content for line 16. For these lines we obtained the best total (calculated for all five traits) genotype selection index (TGSI = 81 and 97 for line 16 and line 7, respectively).
Conclusions
The interspecific cross-derived Brassica lines have not been extensively used in breeding programs because of their low seed yield, low oil content and not-canola quality. However, such plant material is essential for plant breeders to produce cultivars that can develop higher level of resistance to biotic and abiotic stresses.
References
Abakemal D, Shimelis H, Derera J (2016) Genotype-by-environment interaction and yield stability of quality protein maize hybrids developed from tropical-highland adapted inbred lines. Euphytica 209:757–769
Anandan A, Sabesan T, Eswaran R, Rajiv G, Muthalagan N, Suresh R (2009) Appraisal of environmental interaction on quality traits of rice by additive main effects and multiplicative interaction analysis. Cereal Res Commun 37(1):131–140
Bocianowski J, Liersch A, Bartkowiak-Broda I (2014) The relationship between different types of markers and glucosinolates content of parental lines of F1 CMS ogura hybrids of winter oilseed rape (Brassica napus L.). Bulg J Agric Sci 20(4):868–876
Bocianowski J, Nowosad K, Liersch A, Popławska W, Łącka A (2018) Genotype-by-environment interaction for seed glucosinolate content in winter oilseed rape (Brassica napus L.) using an additive main effects and multiplicative interaction model. Biom Lett 55(1):85–96
Chen B, Xu K, Li X, Gao G, Yan G, Qiao J, Wu X (2017) Evaluation of quality traits and their genetic variation in global collections of Brassica napus L. Plant Genet Resour 16(2):146–155
Eckes AH, Annemarie H, Gubała T, Nowakowski P, Szymczyszyn T, Wells R, Irwin JA, Horro C, Hancock JM, King G, Dyer SC, Jurkowski W (2017) Introducing the Brassica information portal: towards integrating genotypic and phenotypic Brassica crop data. F1000Research 6:465. PMC. Web. 9 Sept 2018
Edwards JW (2016) Genotype × environment interaction for plant density response in maize (Zea mays L.). Crop Sci 56:1493–1505
Farshadfar E, Sutka J (2003) Locating QTLs controlling adaptation in wheat using AMMI model. Cereal Res Commun 31:249–256
Fox PN, Crossa J, Ramagosa I (1997) Multienvironment testing and genotype × environment interaction. In: Kempton RA, Fox PN (eds) Statistical methods for plant variety evaluation. Chapman & Hall, London, pp 117–138
Friedt W, Snowdon RJ (2010) Oilseed rape. In: Vollmann J, Rajcan I (eds) Oil crops. Handbook of plant breeding, vol 4. Springer, New York, pp 91–126
Gauch HG, Zobel RW (1990) Imputing missing yield trial data. Theor Appl Genet 79:753–761
Golkari S, Haghparast R, Roohi E, Mobasser S, Ahmadi MM, Soleimani K, Khalilzadeh G, Abedi-Asl G, Babami T (2016) Multi-environment evaluation of winter bread wheat genotypes under rainfed conditions of Iran-using AMMI model. Crop Breed J 4, 5 and 6(2; 1 and 2):17–31
Körber N, Bus A, Li J, Parkin IAP, Wittkop B, Snowdon RJ, Stich B (2016) Agronomic and seed quality traits dissected by genome-wide association mapping in Brassica napus. Front Plant Sci 7:386. https://doi.org/10.3389/fpls.2016.00386
Liersch A, Bocianowski J, Woś H, Szała L, Sosnowska K, Cegielska-Taras T, Nowosad K, Bartkowiak-Broda I (2016) Assessment of genetic relationships in breeding lines and cultivars of Brassica napus and their implications for breeding winter oilseed rape. Crop Sci 56(4):1540–1549
Niemann J, Wojciechowski A, Janowicz J (2012) Broadening the variability of quality traits in rapeseed through interspecific hybridization with an application of immature embryo culture. J Biotechnol Comput Biol Bionanotechnol 93(2):109–115
Nowosad K, Liersch A, Popławska W, Bocianowski J (2016) Genotype by environment interaction for seed yield in rapeseed (Brassica napus L.) using additive main effects and multiplicative interaction model. Euphytica 208:187–194
Nowosad K, Liersch A, Poplawska W, Bocianowski J (2017) Genotype by environment interaction for oil content in winter oilseed rape (Brassica napus L.) using additive main effects and multiplicative interaction model. Indian J Genet Pl Br 77(2):293–297
Purchase JL, Hatting H, van Deventer CS (2000) Genotype × environment interaction of winter wheat (Triticum aestivum L.) in South Africa: II. Stability analysis of yield performance. S Afr J Plant Soil 17:101–107
Sosnowska K, Cegielska-Taras T, Liersch A, Karłowski MW, Bocianowski J, Szała L, Mikołajczyk K, Popławska W (2017) Genetic relationships among resynthesized, semi-resynthesized and natural Brassica napus L. genotypes. Euphytica 213:212
Szała L, Sosnowska K, Popławska W, Liersch A, Olejnik A, Kozłowska K, Bocianowski J, Cegielska-Taras T (2016) Development of new restorer lines for CMS ogura system with the use of resynthesized oilseed rape (Brassica napus L.). Breed Sci 66:516–521
Velasco L, Möllers C, Becker HC (1999) Estimation of seed weight, oil content and fatty acid composition in intact single seeds of rapeseed (Brassica napus L.) by near infrared reflectance spectroscopy. Euphytica 106:79–85
Yan W, Hunt LA (2001) Interpretation of genotype × environment interaction for winter wheat iIn Ontario. Crop Sci 41:19–25
Zobel RW, Wright MJ, Gauch HG (1988) Statistical analysis of yield trial. Agron J 80:388–393
Zou J, Zhao Y, Liu P, Shi L, Wang X, Wang M, Meng J, Reif JC (2016) Seed quality traits can be predicted with high accuracy in Brassica napus using genomic data. PLoS ONE 11(11):e0166624
Acknowledgements
This study was funded by the Polish Ministry of Agriculture and Rural Development.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Bocianowski, J., Niemann, J. & Nowosad, K. Genotype-by-environment interaction for seed quality traits in interspecific cross-derived Brassica lines using additive main effects and multiplicative interaction model. Euphytica 215, 7 (2019). https://doi.org/10.1007/s10681-018-2328-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-018-2328-7