Abstract
Each species is characterized by a specific set of chromosomes, which is described as the chromosome portrait or karyotype. In general, such a karyotype is the same for all individuals in the population. An exception to that rule has recently been found in the orchid Erycina pusilla, which has been reported to have two cytotypes with chromosome numbers of 2n = 10 and 2n = 12. Here, we examined the karyotypes of the two cytotypes and found differences in arm ratios and heterochromatin patterns as well as in the presence of satellite chromosomes and in the number and location of rDNA and telomeric repeat sites. These differences are extensive and would have required multiple chromosome rearrangements to generate the differences between the two karyotypes. We also found that F1 hybrids between the parents with the two different chromosome numbers resulted in sterile offspring, in accordance with our previous findings. The combination of hybrid sterility and extensively rearranged chromosomes supports the hypothesis that these two reported cytotypes are, in fact, two different species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genome of any eukaryotic organism has a characteristic karyotype, with a specific chromosome number and morphology. Such a karyotype is, apart from sex chromosomes and B chromosomes, a constant feature of all individuals of a species. Cytotypes are individuals or populations with one or a few structural or numerical differences in their chromosome sets. Heterozygotes of two different cytotypes can display erratic meiotic behavior, leading to reproductive isolation under certain circumstances, followed by extinction or speciation. The most common cytotypes in a species are those with different chromosome numbers, such as polyploids that have resulted from whole-genome duplications. Polyploidy has been shown to be a common event in the evolution of most angiosperms (De Bodt et al. 2005; Soltis et al. 2008; Jiao et al. 2011; Weiss-Schneeweiss et al. 2013). Polyploids are assumed to have aided survival for land species during and/or after the Cretaceous-Tertiary mass extinction (Fawcett et al. 2009). Cytotypes may also differ in small chromosome number variations, which are caused by aneuploidy or dysploidy. Unlike polyploidy or aneuploidy, dysploidy always involves chromosome structural variation caused by chromosome rearrangements, such as translocations and inversions. Compared to polyploid cytotypes there are far fewer examples of cytotypes with chromosome number polymorphisms caused by dysploidy. Examples are Graomys griseoflavus (2n = 36–38; Cricetidae) (Theiler et al. 1999) and Crocus pallasii subsp. pallasii (2n = 14 and 16; Iridaceae) (Karamplianis et al. 2013). Comparable chromosome polymorphisms have been described in different species of the orchid family, such as Cephalanthera (Moscone et al. 2007) and Paphiopedilum (Lan and Albert 2011). This variability plays a critical role in species evolution and has been considered to be a consequence of chromosome rearrangement (Faria and Navarro 2010; Heslop-Harrison and Schwarzacher 2011). Such rearrangement may increase or decrease the chromosome number in the karyotype and is termed ascending or descending dysploidy (Schubert and Lysak 2011; Leitch et al. 2013; De Storme and Mason 2014; Yang et al. 2014).
Two cytotypes have been reported for Erycina pusilla, i.e., one with 2n = 10 (Dodson 1957a, b) and one with 2n = 12 (Félix and Guerra 1999, 2000). Erycina pusilla (synonym Psygmorchis pusilla) is a member of the orchid family and is known as a type of “twig epiphyte”, which refers to its characteristic of small branch habitat (less than 2.5 cm in diameter) (Kolanowska and Szlachetko 2014). Plants grow naturally under a disturbed environment with low levels of sunlight and irregular water supply, leading to the development of fast-growing and short-juvenile features (Chiu et al. 2011). Unlike most orchid species, E. pusilla often reaches maturity and the flowering stage within 1 year. Accompanied by a smaller genome size (1C = 1.5 pg) (Chase et al. 2005) and low diploid chromosome number (2n = 10 and 12), this tiny orchid (less than 10 cm in height at maturity) is a potential orchid model that has been studied intensely in recent years in such areas as in vitro fruiting and seed production (Chiu et al. 2011), categorization of miRNA (Lin et al. 2013), transcriptome analysis (Chou et al. 2013), chloroplast genome analysis (Pan et al. 2012), establishment of an agrobacterium-mediated genetic transformation procedure (Lee et al. 2015) and transcriptome‐wide analysis of the MADS‐box gene family (Lin et al. 2016). However, little cytogenetic information has been reported: only a chromosome count and conventional karyotype analysis has been carried out in this orchid. In Yeh et al. (2015) the two 2n = 10 and 2n = 12 cytotypes, collected from two South American countries, demonstrated high similarity both in genomic composition and DNA amount based on the results of genomic in situ hybridization and flow cytometry. In addition, analyses of meiotic prophase of the 2n = 10/12 heterozygotes from reciprocal crosses revealed incomplete pairing, with unpaired loops between parental homoeologous chromosome segments. These results suggested that the chromosome number variation of the two cytotypes have resulted from chromosome rearrangements, rather than changes in repeat content of their genomes. However, the establishment and mechanisms of such structural rearrangements in the karyotypes between these two cytotypes remain unclear.
Identification of chromosome rearrangement and karyotype characterization is generally done by chromosome banding or painting methods. However, conventional banding techniques are limited in resolution only 4–6 Mbp chromosome segments, which covers parts of the condensed metaphase complements (de Jong et al. 1999). The Fluorescent in situ Hybridization (FISH) technique based on probes with single copy and repetitive sequences enables detection of much smaller DNA targets, and can provide unique and consistent information for karyotype comparisons for evolutionary and cytotaxonomic analyses. The most common repeats for such studies, for example in Maxillaria (Orchidaceae) (Cabral et al. 2006), Aurantioideae (Moraes et al. 2012), Lupinus (Hajdera et al. 2003), Pinus (Hizume et al. 2002) and Nicotiana (Chen et al. 2002), include ribosomal DNA (45S and 5S rDNA) and telomere repeat sequences (TTTAGGG). The 45S (5.8S–18S–26S) rDNA genes comprise tandemly repetitive units that are highly conserved among species, which are in most species present in the distal parts of the short arms of one or more chromosome pairs called nucleolar organizer regions (NORs). 5S rDNA genes usually localize separately from the 45S rDNA genes and do not take part in nucleolus formation. The telomere repeat sequence is a conserved marker that identifies chromosome ends; however, some studies have revealed interstitial telomere sites (Uchida et al. 2002a, b; Tek and Jiang 2004; Shibata et al. 2005), providing strong evidence of recent chromosome fusions and Robertsonian translocations. In this study, we applied the above-mentioned molecular chromosome markers combined with FISH to determine the karyotype differences between the two cytotypes of E. pusilla.
Materials and methods
Plant material
The two cytotypes of E. pusilla were collected from Ecuador (2n = 10) and from Surinam (2n = 12) and their specimens were deposited in the herbarium of Taiwan’s National Museum of Natural Science (Yeh et al. 2015). All plants were maintained and cultured on half-strength Murashige and Skoog medium (Murashige and Skoog 1962), and cultivated in growth chambers under long-day conditions (16-h photoperiod) at 22 °C.
Chromosome slide preparation
The experimental protocol was according to Chang (Chang et al. 2007, 2008; Szinay et al. 2008) with minor modifications. Fast growing root tips were collected and pre-treated with 2 mM aqueous solution of 8-hydroxyquinoline for 3 h at 15 °C or 1:1 (v/v) 2 mM 8-hydroxyquinoline/0.1% colchicine for 2 h at 15 °C. Root tips were then fixed in freshly prepared Carnoy’s solution (3:1 (v/v) 95% ethanol/glacial acetic acid) and stored at −20 °C until use. The fixed material was washed in distilled water and digested with a pectolytic enzyme mixture containing 1% (w/v) cellulose Onozuka RS (Yakult Honsha), 1% (w/v) pectolyase Y23, and 1% (w/v) pectinase solution) in a humid chamber at 37 °C for 80 min. Subsequently, the fragile meristem was rinsed carefully with distilled water, squashed in a drop of 45% acetic acid, and the coverslip was removed after freezing in liquid nitrogen. The slides were dried at 42 °C and rinsed in Carnoy’s solution and 95% ethanol, respectively. Finally, all slides were observed under a phase-contrast microscope to check the mitotic stage and chromosome quality.
DNA probe labeling
The 9-kb fragment of the 45S rDNA sequences were from Triticum aestivum in plasmid pTA71, and the 5S rDNA sequences were amplified from the genomic DNA of E. pusilla by PCR using the oligomer primer set: 5′-GATCCCATCAGAACTTC-3′ (forward) and 5′-GGTGCTTTAGTGCTGGTAT-3′ (reverse) (Kim et al. 2009). The clone pAt T4 with the Arabidopsis-like telomere repeat (TTTAGGG) was used to detect distal and interstitial telomere sites. All DNA probes were generated by nick translation reaction, following the manufacturer’s instructions (Roche). DNA samples were randomly nicked and labeled with either biotin-16-dUTP or digoxigenin-11-dUTP. Probes were purified by ethanol precipitation and stored at −20 °C.
Fluorescence in situ hybridization (FISH)
Selected high quality squashed root tip slides were placed in a Coplin jar and pretreated with pepsin (5 µg/mL in 0.1 M HCl) at 37 °C for 10 min, 4% formaldehyde solution for 10 min, and a graded series of ethanol (70, 90, 100%) for 3 min each. The hybridization mix (20 µL/slide), containing 50 ng of labeled probes, 50% formamide, 10% sodium dextran sulfate, 0.25% SDS (sodium dodecyl sulfate) and 2 × SSC (saline-sodium citrate) was heated in a boiling water bath for 10 min and then cooled down on ice for 10 min. For the hybridization experiments, probe mix was dropped onto each chromosome slide and covered with a coverslip. Slides were placed on a heating block at 80 °C for 2.5 min and hybridization was carried out in a humid chamber at 37 °C for 16–20 h. After hybridization, coverslips were removed and washed by 2 × SSC twice. Slides were washed three times for 5 min in 50% formamide at 42 °C. Blocking solution (5% bovine serum albumin, 0.2% Tween-20 in 4 × SSC) was added and incubated at 37 °C for 30 min. Subsequently, antibodies (labeled with streptavidin or anti-DIG conjugated fluorescent dye) were added and incubated at 37 °C for 1 h. Slides were washed with 2 × SSC, dehydrated through a graded ethanol series, and mounted with 12 µL of 5 µg/mL of 4′,6-diamidino-2-phenylindole (DAPI) in Vectashield (Vector Laboratories, Burlingame, CA, USA). The chromosome structure and fluorescent signals were observed and photographed using a fluorescence microscope (Zeiss) equipped with a digital camera. Images were pseudo-colored and adjusted using AxioVision software (Zeiss).
Karyotype analysis
At least five metaphase cells of each cytotype, after FISH and DAPI counterstaining, were used for chromosome analysis. Every chromosome was straightened and measured using ImageJ software (http://imagej.nih.gov/ij/) using the plugin of Kocsis et al. (1991). Ideograms were generated according to relative chromosome length, arm ratio (length of long arm/length of short arm), heterochromatin distribution, and location of repetitive sequences (5S and 45S rDNA loci, and Arabidopsis-like telomere repeat sequences). Chromosomes were numbered and arranged in sequence of decreasing length.
Results
Karyotype comparison by DAPI staining
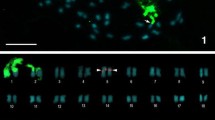
Figure 1 shows representative examples of mitotic metaphase complements of the 2n = 10 and 2n = 12 cytotypes of E. pusilla. Using the chromosome banding as produced by DAPI staining we made ideograms of the two cytotypes. Each chromosome contained a pericentromeric heterochromatin banding pattern and was asymmetrical, with arm ratios ranging from 1.62 to 1.06 (Table 1). The longest pair showed similar arm ratios and heterochromatin patterns in both cytotypes. One chromosome pair of the 2n = 12 cytotype displays secondary constrictions, separating the adjacent satellite from the short arm heterochromatin.
Somatic metaphase chromosomes stained with DAPI and ideograms showing comparative chromosome lengths (arm ratio) with distributions of euchromatic (white) heterochromatic (black) DNA. a Erycina pusilla, 2n = 10. b Erycina pusilla, 2n = 12. Arrows indicate satellite chromosomes (SAT chromosomes). The bar represents 10 µm
Cytological mapping of 45S and 5S rDNA genes
A pair of 45S and three pairs of 5S rDNA loci were located on the 2n = 10 cytotype (Fig. 2a-c). The 45S rDNA signals were on the terminal sites of a homologous chromosome. One pair of 5S signals was present interstitial on one chromosome pair. The other two 5S signals were situated on both ends of another homologous pair, where the 45S rDNA genes were co-localized with 5S rDNA genes at the terminal ends of the long arms. In the 2n = 12 cytotype, one pair of 45S and 5S rDNA loci, were mapped on the distal and interstitial regions of two different homologous chromosomes (Fig. 2d–f), respectively. Thus, the number and location of the 5S rDNA loci differed between these two cytotypes.
Chromosome positions of the telomere sequences
Telomere foci were observed on both ends of each chromosome (Fig. 3a, b). In addition, in the 2n = 12 cytotype, there were four obvious signals located in the centromere regions (Fig. 3b, arrows). Combining conventional (DAPI counterstaining) and molecular cytogenetic data, we constructed ideograms of the two cytotypes (Fig. 4) according to relative chromosome length, heterochromatin banding, satellite structure, arm ratio, and location of fluorescent markers (rDNA and telomere repeat sequence). In addition, Table 2 summarizes the number of rDNA loci, telomere repeat signals, and satellite (SAT) chromosomes between the two cytotypes.
Ideograms of the two cytotypes (different species) showing relative chromosome length, arm ratio, and physical mapping of three molecular makers using FISH techniques: 5S rDNA loci (green), 45S rDNA loci (red), Arabidopsis-like telomeric repeat sequences (orange), and rDNA co-localization (5S + 45S rDNA, yellow). The chromosome pairs are arranged in line with by decreasing length. The numbering of each chromosome is presented according to the decreasing length of each homologous pair of each cytotype/species, respectively
Discussion
We performed FISH and found strikingly different karyotypes between the two cytotypes, based on the different distributions of rDNA, telomere repeat sequences, arm ratio, heterochromatin banding, and satellite location. Furthermore, the karyotypes of each cytotype could provide a valuable reference for chromosome evolution and karyotype analysis in orchids.
Karyotype characterization after DAPI staining
In both cytotypes, the heterochromatic bands were distributed adjacent to primary constrictions with varying quantities of fluorescence. Integrating the banding patterns with chromosome length and arm ratio allowed us to generate ideograms. There is only one chromosome (the longest) that seems to be morphologically identical in both cytotypes. All of the other chromosomes differ in arm ratio, fraction and/or location of heterochromatin, or presence of a satellite. There was one chromosome pair (No. 2) of cytotype 2n = 12 with a secondary constriction. Such constrictions are usually sites of the Nucleolar Organizer Regions (NOR) that provide an essential function in nucleolus formation and function after cell division (Heslop-Harrison and Schwarzacher 2011; Falistocco and Marconi 2013). The NOR is typically the major site of the 45S rDNA gene, but physical mapping of the 45S rDNA loci showed them to be present on chromosome pair No. 5, rather than No. 2. This might have been caused by translocation between these two chromosomes, leading to the relocation of the 45S rDNA genes. Alternatively, the secondary constrictions could be merely a structural feature, not the location of the NOR (Sato et al. 1980). A previous study on Allium sativum indicated that not all NORs are associated with the morphological appearance of secondary constrictions. Another cytological study implied that both the translocation and deletion could be concerned in numerical variation and the polymorphism of the satellite chromosomes of Allium cepa (Sato 1981).
Cytogenetic characterization of the rDNA genes and telomeric repeat sequences
Only one pair of terminal 45S rDNA loci was observed in both the 2n = 10 and 2n = 12 cytotypes. However, the 5S rDNA patterns differed in the two cytotypes. In Fig. 2b, the 2n = 10 cytotype showed three pairs of 5S rDNA signals. One pair of 5S rDNA signals was located interstitially in both cytotypes, similar to the pattern observed in Rhynchospora (Sousa et al. 2011). The other two pairs were located on the same homologous chromosome at the terminus of each arm. This type of arrangement is not observed frequently, but has been found in Cephalanthera longifolia (Moscone et al. 2007), suggesting it could be related to chromosome inversion, with breakages of the 5S rDNA cluster in subtelomere regions. The divergent number and physical location of the 5S rDNA genes could be caused not only by chromosome rearrangement, including inversions, duplications, deletions and translocations (Hanson et al. 1996; Thomas et al. 1996; Raskina et al. 2008; Wang et al. 2013), but also by transposon-mediated transposition events (Raskina et al. 2004; Altinkut et al. 2006; Datson and Murray 2006). Several cases of 5S rDNA-derived sequences being found within transposable elements have been reported (Kapitonov and Jurka 2003; Kalendar et al. 2008). By contrast, loss of rDNA loci during evolution has occurred in Brassicaceae and Rosaceae (Mishima et al. 2002; Hasterok et al. 2006; Liu and Davis 2011). However, the exact mechanism and biological consequence of the variable number of 5S rDNA loci between these two cytotypes remain unknown.
Notably, the 5S rDNA genes were co-localized with the 45S rDNA genes at the long terminal regions of chromosome 5 in cytotype 2n = 10. Such co-localization (adjacent sites) has been reported in studies of the subfamily Aurantioideae (e Silva et al. 2013) and the orchid genus Paphiopedilum (Lan and Albert 2011). In Aurantioideae, the closely linked rDNAs were regarded as an ancestral circumstance and the positional changes of the rDNA linkage might have been caused by chromosome rearrangements or transposition. Furthermore, the adjacent rDNA sites at the terminal ends might be associated with the stabilization of centromere fission events (Dobigny et al. 2003; Chang et al. 2009).
Telomere repeat sequences were located on the ends of every chromosome. Surprisingly, there were two pairs of telomere signals at the centromere regions in the 2n = 12 cytotype. We expected that telomere signals should be found at centromeres or interstitial regions of chromosomes in the 2n = 10 cytotype, because a reduction in chromosome number results commonly from chromosome fusion. However, 2n = 10 possesses a lower chromosome number but has no telomere signals within centromere or interstitial regions. A similar event was documented in the genus Cephalanthera (Moscone et al. 2007), where the species C. damasonium (2n = 36) contained centromeric telomeric signals whereas C. longifolia (2n = 32) did not. The authors suggested the centromere telomere repeats could have been eliminated after Robertsonian fusion in the lower chromosome number species or that telomere signals in the higher chromosome species were not generated by chromosome fusion, and probably occurred through double-stranded DNA breaks associated with telomerase repair or integration of mobile elements. The rDNA loci and telomere patterns between these two cytotypes indicated that E. pusilla has been subjected to complicated chromosome rearrangements during evolution.
Two cytotypes or different species?
Conventional staining techniques combined with molecular markers revealed different karyotype characters in two reported cytotypes of E. pusilla. The variability of chromosome number and genome organization (heterochromatin and rDNA localization) can provide reliable information to analyze the taxonomic relationship (Félix and Guerra 2000, 2010; Koehler et al. 2008). The elevation of chromosome number has been discovered in many orchid genera, generated from the basic chromosome number by hybridization, polyploidization or dysploid formation (Cox et al. 1998; Félix and Guerra 2005). For example, the basic chromosome number of Paphiopedilum was proposed as 2n = 26, and the chromosome number variation under the genus ranged from 2n = 26–42 caused by a Robertsonian centric fission mechanism (Jones 1998; Lee et al. 2011). It has been proposed that the change in chromosome number is from 12 to 10, even initially from 14, in this species (Yeh et al. 2015) because E. pusilla is in the Cymbidioid group, in which the most probable basic chromosome number is x = 7 (Félix and Guerra 2000, 2005, 2010). In this study, we have demonstrated divergent chromosome profiles between two karyotypes that are mostly likely due to multiple steps in chromosome rearrangements. The cytotype/species with 2n = 12 showed comparatively dramatic chromosome differences compared with the cytotype/species with 2n = 10, such as multiple steps with a reduction or duplication in 5S rDNA loci, integration of telomeric repeat sequences in centromere regions, and the formation of a secondary constriction structure (Table 2). In addition, we found a chromosome rearrangement that resulted in co-localization of 5S rDNA and 45S rDNA in the 2n = 12 cytotype. Furthermore, the arm ratios and heterochromatin patterns are also different in some chromosomes except for chromosome 1 s from both cytotypes/species and between chromosome 6 from 2n = 12 and chromosome 3 from 2n = 10, which are more stable with least degree of rearrangement. The different arm ratios and heterochromatin patterns are combinations of chromosome segment translocations and the heterochromatin expansion or deletion events. There is a series of multiple chromosome rearrangements required to generate the differences between the two karyotypes. Our previous result from Yeh et al. (2015) as well as our subsequent efforts also showed no evidence so far for fertile F1 hybrids between the parents with the two different karyotypes. The sterile F1 hybrids and our findings in the study support the hypothesis that these are two separate species, not just cytotypes.
To understand the different karyotypes in details and evolutionary relationship between these two species, more chromosome markers and comparative genetics maps should be generated. This study has demonstrated that cytogenetic work is essential to identify species and the karyotype information also provided a preliminary framework for genome sequencing, genetic map construction, and determining the chromosome evolutionary machinery in these potential orchid model plants.
References
Altinkut A, Raskina O, Nevo E, Belyayev A (2006) En/spm-like transposons in Poaceae species: Transposase sequence variability and chromosomal distribution. Cell Mol Biol Lett 11:214–229
Cabral JS, Félix LP, Guerra M (2006) Heterochromatin diversity and its co-localization with 5S and 45S rDNA sites in chromosomes of four Maxillaria species (Orchidaceae). Genet Mol Biol 29:659–664
Chang SB, Anderson LK, Sherman JD, Royer SM, Stack SM (2007) Predicting and testing physical locations of genetically mapped loci on tomato pachytene chromosome 1. Genetics 176:2131–2138
Chang SB, Yang TJ, Datema E, van Vugt J, Vosman B, Kuipers A, Meznikova M, Szinay D, Lankhorst RK, Jacobsen E, de Jong H (2008) FISH mapping and molecular organization of the major repetitive sequences of tomato. Chromosome Res 16:919–933
Chang Y-C, Shii C-T, Chung M-C (2009) Variations in ribosomal RNA gene loci in spider lily (Lycoris spp.). J Am Soc Hortic Sci 134:567–573
Chase MW, Hanson L, Albert VA, Whitten WM, Williams NH (2005) Life history evolution and genome size in subtribe Oncidiinae (Orchidaceae). Ann Bot 95:191–199
Chen CC, Chen SK, Liu MC, Kao YY (2002) Mapping of DNA markers to arms and sub-arm regions of Nicotiana sylvestris chromosomes using aberrant alien addition lines. Theor Appl Genet 105:8–15
Chiu YT, Lin CS, Chang C (2011) In vitro fruiting and seed production in Erycina Pusilla (L.) N. H. Williams & M. W. Chase. Propag Ornam Plants 11:131–136
Chou ML, Shih MC, Chan MT, Liao SY, Hsu CT, Haung YT, Chen JJW, Liao DC, Wu FH, Lin CS (2013) Global transcriptome analysis and identification of a CONSTANS-like gene family in the orchid Erycina pusilla. Planta 237:1425–1441
Cox AV, Abdelnour GJ, Bennett MD, Leitch IJ (1998) Genome size and karyotype evolution in the slipper orchids (Cypripedioideae : Orchidaceae). Am J Bot 85:681–687
Datson PM, Murray BG (2006) Ribosomal DNA locus evolution in Nemesia: transposition rather than structural rearrangement as the key mechanism? Chromosome Res 14:845–857
de Bodt S, Maere S, Van de Peer Y (2005) Genome duplication and the origin of angiosperms. Trends Ecol Evol 20:591–597
de Jong JH, Fransz P, Zable P (1999) High resolution FISH in plants-techniques and applications. Trends Plant Sci 4:258–263
de Storme N, Mason A (2014) Plant speciation through chromosome instability and ploidy change: Cellular mechanisms, molecular factors and evolutionary relevance. Curr Plant Biol 1:10–33
Dobigny G, Ozouf-costaz C, Bonillo C, Volobouev V (2003) Evolution of rRNA gene clusters and telomeric repeats during explosive genome repatterning in Taterillus X (Rodentia, Gerbillinae). Cytogenet Genome Res 103:94–103
Dodson C (1957a) Chromosome number in Oncidium and allied genera. Am Orchid Soc Bull 26:323–330
Dodson C (1957b) Oncidium pusillum and its allies I. Am Orchid Soc Bull 26:170–172
e Silva AB, Barros AE, Dos Santos Soares Filho W, Guerra M (2013) Linked 5S and 45S rDNA sites are highly conserved through the subfamily Aurantioideae (Rutaceae). Cytogenet Genome Res 140:62–69
Falistocco E, Marconi G (2013) Cytogenetic characterization by in situ hybridization techniques and molecular analysis of 5S rRNA genes of the European hazelnut (Corylus avellana). Genome 56:155–159
Faria R, Navarro A (2010) Chromosomal speciation revisited: rearranging theory with pieces of evidence. Trends Ecol Evol 25:660–669
Fawcett JA, Maere S, Van de Peer Y (2009) Plants with double genomes might have had a better chance to survive the Cretaceous-Tertiary extinction event. Proc Natl Acad Sci USA 106:5737–5742
Félix LP, Guerra M (1999) Chromosome analysis in Psygmorchis pusilla (L.) Dodson & Dressier: the smallest chromosome number known in Orchidaceae. Caryologia 52:165–168
Félix LP, Guerra M (2000) Cytogenetics and cytotaxonomy of some Brazilian species of Cymbidioid orchids. Genet Mol Biol 23:957–978
Félix LP, Guerra M (2005) Basic chromosome numbers of terrestrial orchids. Plant Syst Evol 254:131–148
Félix LP, Guerra M (2010) Variation in chromosome number and the basic number of subfamily Epidendroideae (Orchidaceae). Bot J Linn Soc 163:234–278
Hajdera I, Siwinska D, Hasterok R, Maluszynska J (2003) Molecular cytogenetic analysis of genome structure in Lupinus angustifolius and Lupinus cosentinii. Theor Appl Genet 107:988–996
Hanson RE, IslamFaridi MN, Percival EA, Crane CF, Ji YF, McKnight TD, Stelly DM, Price HJ (1996) Distribution of 5S and 18S-28S rDNA loci in a tetraploid cotton (Gossypium hirsutum L) and its putative diploid ancestors. Chromosoma 105:55–61
Hasterok R, Wolny E, Hosiawa M, Kowalczyk M, Kulak-Ksiazczyk S, Ksiazczyk T, Heneen WK, Maluszynska J (2006) Comparative analysis of rDNA distribution in chromosomes of various species of Brassicaceae. Ann Bot 97:205–216
Heslop-Harrison JS, Schwarzacher T (2011) Organisation of the plant genome in chromosomes. Plant J 66:18–33
Hizume M, Shibata F, Matsusaki Y, Garajova Z (2002) Chromosome identification and comparative karyotypic analyses of four Pinus species. Theor Appl Genet 105:491–497
Jiao Y, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, Tomsho LP, Hu Y, Liang H, Soltis PS, Soltis DE, Clifton SW, Schlarbaum SE, Schuster SC, Ma H, Leebens-Mack J, de Pamphilis CW (2011) Ancestral polyploidy in seed plants and angiosperms. Nature 473:97–100
Jones K (1998) Robertsonian fusion and centric fission in karyotype evolution of higher plants. Bot Rev 64:273–289
Kalendar R, Tanskanen J, Chang W, Antonius K, Sela H, Peleg O, Schulman AH (2008) Cassandra retrotransposons carry independently transcribed 5S RNA. Proc Natl Acad Sci USA 105:5833–5838
Kapitonov VV, Jurka J (2003) A novel class of SINE elements derived from 5S rRNA. Mol Biol Evol 20:694–702
Karamplianis T, Tsiftsis S, Constantinidis T (2013) The genus Crocus (Iridaceae) in Greece: some noteworthy floristic records and karyotypes. Phytol Balc 19:1
Kim H, Choi SR, Bae J, Hong CP, Lee SY, Hossain MJ, Van Nguyen D, Jin M, Park BS, Bang JW, Bancroft I, Lim YP (2009) Sequenced BAC anchored reference genetic map that reconciles the ten individual chromosomes of Brassica rapa. BMC Genom 10:432
Kocsis E, Trus BL, Steer CJ, Bisher ME, Steven AC (1991) Image averaging of flexible fibrous macromolecules: the clathrin triskelion has an elastic proximal segment. J Struct Biol 107:6–14
Koehler S, Cabral JS, Whitten WM, Williams NH, Singer RB, Neubig KM, Guerra M, Souza AP, Amaral MDCE (2008) Molecular phylogeny of the neotropical genus Christensonella (Orchidaceae, Maxillariinae): species delimitation and insights into chromosome evolution. Ann Bot 102:491–507
Kolanowska M, Szlachetko DL (2014) Notes on Erycina-complex with descriptions of new Colombian species. Plant Syst Evol 300:527–534
Lan TY, Albert VA (2011) Dynamic distribution patterns of ribosomal DNA and chromosomal evolution in Paphiopedilum, a lady’s slipper orchid. BMC Plant Biol 11:15
Lee YI, Chang FC, Chung MC (2011) Chromosome pairing affinities in interspecific hybrids reflect phylogenetic distances among lady’s slipper orchids (Paphiopedilum). Ann Bot 108:113–121
Lee SH, Li CW, Liau CH, Chang PY, Liao LJ, Lin CS, Chan MT (2015) Establishment of an Agrobacterium-mediated genetic transformation procedure for the experimental model orchid Erycina pusilla. Plant Cell Tissue Organ Cult 120:211–220
Leitch IJ, Greilhuber J, Dolezel J, Wendel JF (2013) Plant genome diversity, vol 2. Springer, Vienna
Lin CS, Chen JJW, Huang YT, Hsu CT, Lu HC, Chou ML, Chen LC, Ou CI, Liao DC, Yeh YY, Chang SB, Shen SC, Wu FH, Shih MC, Chan MT (2013) Catalog of Erycina pusilla miRNA and categorization of reproductive phase-related miRNAs and their target gene families. Plant Mol Biol 82:193–204
Lin CS, Hsu CT, Liao DC, Chang WJ, Chou ML, Huang YT, Chen JJW, Ko SS, Chan MT, Shih MC (2016) Transcriptome-wide analysis of the MADS-box gene family in the orchid Erycina pusilla. Plant Biotechnol J 14:284–298
Liu B, Davis TM (2011) Conservation and loss of ribosomal RNA gene sites in diploid and polyploid Fragaria (Rosaceae). BMC Plant Biol 11:157
Mishima M, Ohmido N, Fukui M, Yahara T (2002) Trends in site-number change of rDNA loci during polyploid evolution in Sanguisorba (Rosaceae). Chromosoma 110:550–558
Moraes AP, Leitch IJ, Leitch AR (2012) Chromosome studies in Orchidaceae: karyotype divergence in Neotropical genera in subtribe Maxillariinae. Bot J Linn Soc 170:29–39
Moscone EA, Samuel R, Schwarzacher T, Schweizer D, Pedrosa-Harand A (2007) Complex rearrangements are involved in Cephalanthera (Orchidaceae) chromosome evolution. Chromosome Res 15:931–943
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15:473–497
Pan IC, Liao DC, Wu FH, Daniell H, Singh ND, Chang C, Shih MC, Chan MT, Lin CS (2012) Complete chloroplast genome sequence of an orchid model plant candidate: Erycina pusilla apply in tropical oncidium breeding. PLoS ONE 7:12
Raskina O, Belyayev A, Nevo E (2004) Activity of the En/Spm-like transposons in meiosis as a base for chromosome repatterning in a small, isolated, peripheral population of Aegilops speltoides Tausch. Chromosome Res 12:153–161
Raskina O, Barber JC, Nevo E, Belyayev A (2008) Repetitive DNA and chromosomal rearrangements: speciation-related events in plant genomes. Cytogenet Genome Res 120:351–357
Sato S (1981) Cytological studies on the satellited chromosomes of Allium cepa. Caryologia 34:431–440
Sato S, Hizume M, Kawamura S (1980) Relationship between secondary constrictions and nucleolus organizing regions in Allium sativum chromosomes. Protoplasma 105:77–85
Schubert I, Lysak MA (2011) Interpretation of karyotype evolution should consider chromosome structural constraints. Trends Genet 27:207–216
Shibata F, Matsusaki Y, Hizume M (2005) AT-rich sequences containing Arabidopsis-type telomere sequence and their chromosomal distribution in Pinus densiflora. Theor Appl Genet 110:1253–1258
Soltis DE, Bell CD, Kim S, Soltis PS (2008) Origin and early evolution of angiosperms. Ann N Y Acad Sci 1133:3–25
Sousa A, e Silva AB, Cuadrado A, Loarce Y, Alves M, Guerra M (2011) Distribution of 5S and 45S rDNA sites in plants with holokinetic chromosomes and the “chromosome field” hypothesis. Micron 42:625–631
Szinay D, Chang SB, Khrustaleva L, Peters S, Schijlen E, Bai YL, Stiekema WJ, van Ham R, de Jong H, Lankhorst RMK (2008) High-resolution chromosome mapping of BACs using multi-colour FISH and pooled-BAC FISH as a backbone for sequencing tomato chromosome 6. Plant J 56:627–637
Tek AL, Jiang JM (2004) The centromeric regions of potato chromosomes contain megabase-sized tandem arrays of telomere-similiar sequence. Chromosoma 113:77–83
Theiler G, Gardenal C, Blanco A (1999) Patterns of evolution in Graomys griseoflavus (Rodentia, Muridae). IV. A case of rapid speciation. J Evol Biol 12:970–979
Thomas HM, Harper JA, Meredith MR, Morgan WG, Thomas ID, Timms E, King IP (1996) Comparison of ribosomal DNA sites in Lolium species by fluorescence in situ hybridization. Chromosome Res 4:486–490
Uchida W, Matsunaga S, Sugiyama R, Kawano S (2002a) Interstitial telomere-like repeats in the Arabidopsis thaliana genome. Genes Genet Syst 77:63–67
Uchida W, Matsunaga S, Sugiyama R, Shibata F, Kazama Y, Miyazawa Y, Hizume M, Kawano S (2002b) Distribution of interstitial telomere-like repeats and their adjacent sequences in a dioecious plant, Silene latifolia. Chromosoma 111:313–320
Wang GX, Li H, Cheng ZK, Jin WW (2013) A novel translocation event leads to a recombinant stable chromosome with interrupted centromeric domains in rice. Chromosoma 122:295–303
Weiss-Schneeweiss H, Emadzade K, Jang TS, Schneeweiss GM (2013) Evolutionary consequences, constraints and potential of polyploidy in plants. Cytogenet Genome Res 140:137–150
Yang L, Koo DH, Li D, Zhang T, Jiang J, Luan F, Renner SS, Henaff E, Sanseverino W, Garcia-Mas J, Casacuberta J, Senalik DA, Simon PW, Chen J, Weng Y (2014) Next-generation sequencing, FISH mapping and synteny-based modeling reveal mechanisms of decreasing dysploidy in Cucumis. Plant J 77:16–30
Yeh HY, Lin CS, Chang SB (2015) Cytogenetic and cytometric analyses in artificial intercytotypic hybrids of the emergent orchid model species Erycina pusilla. Euphytica 206:533–539
Acknowledgements
The Ministry of Science and Technology (MOST), Taiwan, R.O.C. supported this research under Grant Nos. 104-2313-B-006-001 and 106-2313-B-006 002.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Yeh, HY., Lin, CS., de Jong, H. et al. Two reported cytotypes of the emergent orchid model species Erycina pusilla are two different species. Euphytica 213, 233 (2017). https://doi.org/10.1007/s10681-017-2026-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-017-2026-x