Abstract
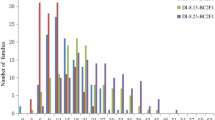
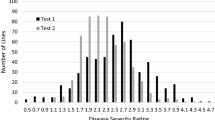
Verticillium wilt (VW), caused by Verticillium dahliae Kleb., is one of the most important diseases in cotton. The objective of this study was to map quantitative trait loci (QTLs) conferring VW resistance using resistance gene analog (RGA)-targeted amplified fragment length polymorphism (RGA-AFLP) markers in an interspecific backcross inbred line mapping population, consisting of 146 lines from a susceptible Sure-Grow 747 (Gossypium hirsutum L.) × resistant Pima S-7 (G. barbadense L.) cross. VW resistance was evaluated in replicated tests based on disease incidence in the field, and disease incidence and severity in the greenhouse. Of 160 polymorphic RGA-AFLP markers, 42 were significantly correlated with one or more VW traits and 41 were placed on a linkage map which covered 1,226 cM of the cotton genome and contained 251 other molecular markers. Three QTLs for VW resistance were detected, each of which explained 12.0–18.6 % of the phenotypic variation. Two of these QTLs for disease incidence and severity detected in the greenhouse inoculation tests using root wounding are located on chromosome c4. Both are closely linked to four RGA-AFLP markers and therefore considered as the same QTL for VW resistance. The other QTL detected in the field test was located on c19 and flanked by several RGA-AFLP markers. The desirable QTL allele on c4 for VW resistance detected in the greenhouse was from the VW susceptible Upland parent and absent from the resistant Pima parent which was more VW susceptible due to the disarmament of the first line of defense mechanism due to root wounding during inoculation. The other desirable VW resistance QTL allele, on c19, was from the resistant parent Pima S-7, consistent with the fact that Pima cotton was more resistant to VW when naturally infected in the field. The results should facilitate the development of more sequence specific markers and the transfer of VW resistance from Pima to Upland cotton through marker-assisted selection.

Similar content being viewed by others
References
Abdelraheem A, Tiwari R, Zhang JF (2012) Genetic analysis and QTL mapping of drought tolerance in cotton under PEG conditions. Proc. Beltwide Cotton Conference, pp 719–728
Adams N, Flynn R, Bajaj S, Percy RG, Jones DC, Hughs SE, Zhang JF (2011) Identification of cotton germplasm and molecular markers for drought tolerance. Proc. Beltwide Cotton Conference, p 708
Bell AA (1969) Phytoalexin production and Verticillium wilt resistance in cotton. Phytopathology 59:1119–1127
Blasingame D, Patel MV (2011) Cotton disease loss estimate committee report, 2011 Beltwide Cotton Conferences, Atlanta, pp 306–308
Blenda A, Fang DD, Rami JM, Garsmeur O, Luo M, Lacape JM (2012) A high density consensus genetic map of tetraploid cotton that integrates multiple component maps through molecular marker redundancy check. PLoS One 7:e45739
Bolek Y, Bell AA, El-Zik KM, Thaxton PM, Magill CW (2005a) Reaction of cotton cultivars and an F2 population to stem inoculation with isolates Verticillium dahliae. J Phytopathol 153:269–273
Bolek Y, El-Zik KM, Pepper AE, Bell AA, Magill CW, Thaxton PM, Reddy OUK (2005b) Mapping of Verticillium wilt resistance genes in cotton. Plant Sci 168:1581–1590
Carpenter CW (1914) The Verticillium wilt problem. Phytopathology 4:393
Chen XM, Line RF, Leung H (1998) Genome scanning for resistance-gene analogs in rice, barley, and wheat by high-resolution electrophoresis. Theor Appl Genet 97:345–355
Eitas TK, Dangl JL (2010) NB-LRR proteins: pairs, pieces, perception, partners, and pathways. Curr Opin Plant Biol 13:472–477
El-Zik KM (1985) Integrated control of Verticillium wilt of cotton. Plant Dis 69:1025–1032
Feuillet C, Schachermayr G, Keller B (1997) Molecular cloning of a new receptor-like kinase gene encoded at the Lr10 disease resistance locus of wheat. Plant J 11:45–52
Fradin EF, Thomma BPHJ (2006) Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo-atrum. Mol Plant Pathol 7:71–86
Garber R, Houston B (1966) Penetration and development of Verticillium albo-atrum in the cotton plant. Phytopathology 56:1121–1126
Guo WZ, Cai CP, Wang CB, Han ZG, Song XL, Wang K, Niu XW, Wang C, Lu KY, Shi B, Zhang TZ (2007) A Microsatellite-based, gene-rich linkage map reveals genome structure, function and evolution in Gossypium. Genetics 176:527–541
Hill MK, Lyon KJ, Lyon BR (1999) Identification of disease response genes expressed in Gossypium hirsutum upon infection with the wilt pathogen Verticillium dahliae. Plant Mol Biol 40:289–296
Hinchliffe D, Lu Y, Potenza C, Segupta-Gopalan C, Cantrell RG, Zhang JF (2005) Resistance gene analogue markers are mapped to homeologous chromosomes in cultivated tetraploid cotton. Theor Appl Genet 110:1074–1085
Kosambi DD (1943) The estimation of map distances from recombination values. Ann Eugen 12:172–175
Lacape JM, Llewellyn D, Jacobs J, Arioli T, Becker D, Calhoun S, Al-Ghazi Y, Liu S, Palai O, Georges S, Giband M, de Assuncao, Barroso PA, Claverie M, Gawryziak G, Jean J, Vialle M, Viot C (2010) Meta-analysis of cotton fiber quality QTLs across diverse environments in a Gossypium hirsutum × G. barbadense RIL population. BMC Plant Biol 10:132
Leister D, Ballvora A, Salamini F, Gebhardt C (1996) A PCR-based approach for isolating pathogen resistance genes from potato with potential for wide application in plants. Nat Genet 14:421–429
Lüders RR, Galbieri R, Fuzatto MG, Cia E (2008) Inheritance of resistance to Verticillium wilt in cotton. Crop Breed Appl Biotechnol 8:265–270
Martin GB (1999) Functional analysis of plant disease resistance genes and their downstream effectors. Curr Opin Plant Biol 2:273–279
Martin GB, Bogdanove AJ, Sessa G (2003) Understanding the functions of plant disease resistance proteins. Annu Rev Plant Biol 54:23–61
McCouch S, Cho Y, Yano M, Paul E, Blinstrub M, Morishima H, Kinoshita T (1997) Report on QTL nomenclature. Rice Genet Newsl 14:11–13
Mutlu N, Miklas PN, Coyne DP (2006) Resistance gene analog polymorphism (RGAP) markers co-localize with disease resistance genes and QTL in common bean. Mol Breed 17:127–135
Niu C, Lu Y, Yuan Y, Percy R, Ulloa M, Zhang JF (2011) Mapping resistance gene analogs (RGAs) in cultivated tetraploid cotton using RGA-AFLP analysis. Euphytica 181:65–76
Pang M (2008) Transcriptome analysis and development of molecular markers for fiber development. Ph.D. dissertation, New Mexico State University, Las Cruces
Pang M, Percy RG, Hughs E, Zhang JF (2009) Promoter anchored amplified polymorphism based on random amplified polymorphic DNA (PAAP-RAPD) in cotton. Euphytica 167:281–291
Pang M, Percy R, Hughs S, Jones D, Zhang JF (2012) Identification of genes that were differentially expressed and associated with fiber yield and quality using cDNA-AFLP and a backcross inbred line population. Mol Breed 30:975–985
Pegg GF, Brady BL (2002) Verticillium wilts. CABI Publishing, Wallingford, pp 1–300
Shen KA, Meyers BC, Islam-Faridi MN, Chin DB, Stelly DM, Michelmore RW (1998) Resistance gene candidates identified by PCR with degenerate oligonucleotide primers map to clusters of resistance genes in lettuce. Mol Plant Microbe Interact 11:815–823
Smith CW, Cothren JT (1999) Cotton: origin, history, technology, and production. Wiley, New York
Tan H, Callahan FE, Zhang XD, Karaca M, Saha S, Jenkins JN, Creech RG, Ma DP (2003) Identification of resistance gene analogs in cotton (Gossypium hirsutum L.). Euphytica 134:1–7
Tanksley SD, Nelson JC (1996) Advanced backcross QTL analysis: a method for the simultaneous discovery and transfer of valuable QTLs from unadapted germplasm into elite breeding lines. Theor Appl Genet 92:191–203
Tiwari R (2012) Identification of molecular markers and quantitative trait loci for salt tolerance in a backcross inbred line population of cotton. Ph.D. dissertation, New Mexico State University, Las Cruces
Tiwari RS, Picchioni G, Steiner RL, Hughs SE, Jones DC, Zhang JF (2013a) Genetic variation in salt tolerance during seed germination in a backcross inbred line population and advanced breeding lines derived from Upland Cotton × Pima cotton. Crop Sci. doi:10.2135/cropsci2013.01.0028
Tiwari RS, Picchioni G, Steiner RL, Hughs SE, Jones DC, Zhang JF (2013b) Genetic variation in salt tolerance at the seedling stage in an interspecific backcross inbred line population of cultivated tetraploid cotton. Euphytica. doi:10.1007/s10681-013-0927-x
Turcotte EL, Percy RG, Feaster RG (1992) Registration of ‘Pima S-7’ American Pima cotton. Crop Sci 32:1291
USDA-NASS (2013) Crop Production 2012 Summary. January 2013. http://usda01.library.cornell.edu/usda/current/CropProdSu/CropProdSu-01-11-2013.pdf. Accessed 17 Mar 2013
van Ooijen JW (2006) JoinMap® 4.0, Software for the calculation of genetic linkage maps in experimental populations. Kyazma B.V. Wageningen, Netherlands
Vos P, Hogers R, Bleeker M, Reijans M, Lee T, Hornes M, Friters A, Pot J, Paleman J, Kuiper M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Wang HM, Lin ZX, Zhang XL, Chen W, Guo XP, Nie YC, Li YH (2008) Mapping and quantitative trait loci analysis of Verticillium wilt resistance genes in cotton. J Integr Plant Biol 50:174–182
Wendel JF, Cronn RC (2003) Polyplody and the evolutionary history of cotton. Adv Agron 78:139–186
Wilhelm S (1955) Longevity of the Verticillium wilt fungus in the laboratory and in the field. Phytopathology 45:180–181
Wilhelm S, Sagen JE, Tietz H (1969) Dominance of Sea Island resistant to Verticillium wilt in F1 progenies of Upland × Sea Island cotton crosses. Proc Beltwide Cotton Prod Res Conf pp.31
Wilhelm S, Sagen JE, Tietz H (1972) Resistance to Verticillium wilt transferred from Gossypium barbadense to Upland cotton phenotype. Phytopathology 62:798–799
Wilhelm S, Sagen JE, Tietz H (1974) Resistance to Verticillium wilt in cotton: sources, techniques of identification, inheritance trends, and resistance potential of multiline cultivars. Phytopathology 64:924–931
Yang C, Guo WZ, Li GY, Gao F, Lin SS, Zhang TZ (2008a) QTLs mapping for Verticillium wilt resistance at seedling and maturity stages in Gossypium barbadense L. Plant Sci 174:290–298
Yang J, Hu CC, Hu H, Yu RD, Xia Z, Ye XZ, Zhu J (2008b) QTLNetwork: mapping and visualizing genetic architecture of complex traits in experimental populations. Bioinformatics 24:721–723. (QTLNetwork-2.1 user manual—software for mapping QTL with epistatic and QE interaction effects: www.ibi.zju.edu.cn/software/qtlnetwork)
Yu YG, Buss GR, Maroof MA (1996) Isolation of a superfamily of candidate disease-resistance genes in soybean based on a conserved nucleotide-binding site. Proc Natl Acad Sci USA 93:11751–11756
Yu JW, Yu SX, Liu C, WuW, Fan SL, Song MZ, Lin ZX, Zhang XL, Zhang JF (2007) High-density linkage map of cultivated allotetraploid cotton based on SSR, TRAP, SRAP and AFLP markers. J Integr Plant Biol 49:716–724
Yu Y, Yuan D, Liang S, Li X, Wang X, Lin Z, Zhang X (2011) Genome structure of cotton revealed by a genome-wide SSR genetic map constructed from a BC1 population between Gossypium hirsutum and G. barbadense. BMC Genomics 12:15
Yu JW, Yu SX, Fan SL, Song MZ, Zhai HH, Li XL, Zhang JF (2012) Mapping quantitative trait loci for cottonseed oil, protein and gossypol content in a Gossypium hirsutum × Gossypium barbadense backcross inbred line population. Euphytica 187:191–201
Yu JW, Zhang K, Li S, Yu S, Zhai H, Wu M, Li X, Fan S, Song M, Yang D, Li Y, Zhang JF (2013) Mapping quantitative trait loci for lint yield and fiber quality across environments in a Gossypium hirsutum × Gossypium barbadense backcross inbred line population. Theor Appl Genet 126:275–287
Zhang JF, Percy RG (2007) Improving Upland cotton by introducing desirable genes from Pima cotton. World Cotton Res Conf. http://wcrc.confex.com/wcrc/2007/techprogram/P1901.HTM. Accessed 16 Apr 2013
Zhang JF, Percy RG (2008) Genetic analysis of yield and fiber quality traits in a backcross inbred line population of cotton. ASA-CSSA-SSSA Ann Meetings, 5–9 Oct 2008, Houston. http://a-c-s.confex.com/crops/2008am/webprogram/Paper42279.html. Accessed 16 Apr 2013
Zhang JF, Stewart JM (2000) Economical and rapid method for extracting cotton genomic DNA. J Cotton Sci 4:193–201
Zhang JF, Lu Y, Adragna H, Hughs E (2005) Genetic improvement of New Mexico Acala cotton germplasm and their genetic diversity. Crop Sci 45:2363–2373
Zhang JF, Yuan YL, Niu C, Hinchliffe DJ, Lu YZ, Yu SX, Percy RG, Ulloa M, Cantrell RG (2007) AFLP-RGA markers in comparison with RGA and AFLP in cultivated tetraploid cotton. Crop Sci 47:180–187
Zhang JF, Sanogo S, Flynn R, Baral JB, Bajaj S, Hughs SE, Percy RG (2012) Germplasm evaluation and transfer of Verticillium wilt resistance from Pima (Gossypium barbadense) to Upland cotton (G. hirsutum). Euphytica 187:147–160
Acknowledgments
The research was in part funded by USDA-ARS, Cotton Incorporated and New Mexico Agricultural Experiment Station.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Fang, H., Zhou, H., Sanogo, S. et al. Quantitative trait locus mapping for Verticillium wilt resistance in a backcross inbred line population of cotton (Gossypium hirsutum × Gossypium barbadense) based on RGA-AFLP analysis. Euphytica 194, 79–91 (2013). https://doi.org/10.1007/s10681-013-0965-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-013-0965-4