Abstract
Heavy metals are considered the most common pollutants in industrial wastewater areas. Out of thirty bacterial isolates, only 3 isolates sighted the highest metal resistance activity for Zn+2, Fe+2, Pb+2, Co+2, Mn+2, Ni+2, and Cd+2. The biochemical and DNA homology identification with similarities 99.58%, 99.79%, and 99.86% of those isolates was identified and deposited in WDCM, respectively, as Enterobacter kobei OM144907 SCUF0000311, Enterobacter cloacae OM180597 SCUF0000312, and Enterobacter hormaechei OM181067 SCUF0000313. The minimum tolerance activity (MIC) of heavy metal concentrations against E. kobei and E. cloacae was 25, 15, and 15 mmol/l for Ni+2, Fe+2, and Mn+2, respectively, and 10 mmol/l for Zn+2, Pb+2, Co+2, and Cd+2, while against E. hormaechei, it is 15 mmol/l for Ni+2, Fe+2, and Mn+2 and 10 mmol/l for Zn+2, Pb+2, Co+2, and Cd+2. The consortium and solitary application of bacterial isolates towards heavy metal removal at 100%, 200%, and 300% industrial wastewater concentrations were conducted and showed that more than 90% removal of Zn+2, Fe+2, Pb+2, Mn+2, Ni+2, and Cd+2 from a non-concentrated polluted sample (100%) was reported by the three strains. With doubling the polluted sample concentration (200%), the highest removal efficiency for Zn+2, Pb+2, Mn+2, Ni+2, and Cd+2 was reported by E. cloacae as 70. 75, 66, 65, and 57%, respectively. Removal efficiency after increasing the polluted sample concentration to 300% showed that E. cloacae removed above 45% of all tested heavy metals except Pb+2. Ultimately, E. cloacae exposed the highest efficiency with recommendations for heavy metals removal under higher concentrations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Organic and inorganic pollutants that enter the marine environment have the worst impact and possess a main hazard to all environments and universal ecosystems. Heavy metals, in particular, act as the most influencing hazardous waste that could harm living organisms in any ecosystem. Such harmfulness refers to its toxicity, bioaccumulation, non-degradability, and bio-amplification through progressive trophic levels (Ayaz et al., 2020). A variety of techniques have been applied for remediating the heavy metal contaminants such as precipitation and membrane technologies in addition to ion exchange and electrochemical processes and eventually the biological methods (Ilavský et al., 2015). Generally, heavy metals in trace amounts are playing as essential elements in many metabolic activities of living organisms; however, beyond a certain threshold, they become toxic elements for those organisms causing varying diseases and unstable behavior in living organisms and their ecological systems concerning the non-degradable characteristic of such elements (Mustapha & Halimoon, 2015). As an emerging technique for heavy metal bioremediation, biosorption has proved to be an efficient approach from a point of view of simplicity, flexibility, efficiency, and low-cost methodology focusing on binding the heavy metals on cellular surface structures of biomasses such as bacteria, yeast, fungi, and algae (Espinosa-Ortiz et al., 2016; Rahman et al., 2019).
Microbes are present in our rounded environment, especially in presence of essential elements for growth, where the pollutants may act as co-factors for bacterial growth within certain thresholds. In a sense of that, industrial waste estuaries are considered a suitable place for adopting the growth of all types of microorganisms with certain limitations. For instance, nickel, iron, cobalt, and zinc, which are the dominant industrial waste, play the growth key factor for many bacterial communities, where they possess the appropriate approach to adopt, uptake, and convert them to its beneficiary target (Figueira et al., 2005). Recently, scientists have tended to use bacteria to remove or reduce heavy metals in water and soil. One of those remarkable bacterial families is Enterobacteriaceae. For instance, Enterobacter sp., Enterobacter cloacae, and Enterobacter asburiae are used for bioremediation of Cu+2, Cr+2, Pb+2, Cd+2, and Ni+2 from different pollution sites (Banerjee et al., 2015; Bestawy et al., 2013; Paul & Mukherjee, 2016; Rahman et al., 2015).
The degree of heavy metal pollution in terms of accumulation pattern is more determined in sediment than in Seawater, where the sediment grain size gives an estimation of the sources, occurrences, and distributions of heavy metals in coastal and estuarine sediments. On the other side, a variety of natural heavy metal accumulation is often located in marine sediments in the shallow and sheltered zones giving the historical variations and the influencing of human activities input in the marine ecosystem (Alloway, 2012; Guagliardi et al., 2013). The retention of heavy metals in marine sediments is probably organized by the rates of finest fractions accumulation, the organic matter decomposition, and Fe+2 and Mn+2 concentrations (Dar et al., 2016). Consequently, the aim of the current research paper was (i) sample collection targeting the isolation of highly potential tolerant microbes, (ii) minimum tolerance activity (MIC) of isolates for different metal concentrations, (iii) identification of most potent isolates, (iv) sediment sieve analysis, and (v) evaluation of solo and consortium potential isolates towards heavy metal removal of different concentrations: 100%, 200%, and 300% of drainage wastewater.
Materials and methods
Sample description and collection
The water and sewage samples were collected under sterilized conditions from different sites of the main industrial estuary drainage in the Adabiya area, Suez, Egypt, in 2021 (supplementary file b figure 1S-a). Samples were aseptically processed for isolation of bacterial spp. using a mineral medium with composition 1 g K2HPO4, 1 g KH2PO4, 0.1 g NaCl, 1 g NH4NO3, 0.5 g MgSO4·7H2O, 0.1 g Pb (CH3COO)4, 0.1 g CuSO4, 0.1 g ZnSO4, 0.1 g Co(NO3)2·6 H2O, 10 g yeast extract, 10 g beef extract, and 0.02 g CaCl2 in 1 L H2O. The neutral pH level of the prepared medium was adjusted to 7 and incubated for 72 h at 37°C. Supplied chemicals of Sigma Aldrich grad were incorporated in the current research. After incubation, the grown separated bacterial cells were isolated and subcultured using the previous mineral agar medium. To generate the bacterial inoculum for bioremediation, all bacterial isolates were cultivated in a nutrient broth at 37 °C with a shaking speed of 130 rpm for 24 h (Ijoma et al., 2019).
Heavy metal resistance assessment
The tolerance test depended on the bacterial growth with and without lead acetate, copper sulfate, zinc sulfate, and cobalt nitrate as a metal supplement for medium and bacterial isolates. Briefly, the 30 bacterial isolates were incubated in nutrient broth, and then each isolate was inoculated in five separate flasks. The first flask did not contain any metal supplement with medium and other flasks contained lead acetate, copper sulfate, zinc sulfate, and cobalt nitrate by 1 mM concentration with medium, respectively (Muñoz et al., 2012). Bacterial cell growth for all flasks was determined by measuring OD at 600 nm and microbial counts as colony-forming units (CFU/mL) by serial dilution method (Verma & Kuila, 2019). On the other hand, the agar diffusion method was used to determine the resistance of bacterial isolates to different heavy metals. Well, diffusion plates were prepared using sterile cork borer with poured nutrient agar plates inoculated with overnight cultures of target strains, where 200 microns μm (200 μl) of known concentration (10mmol/l) of tested heavy metals solutions were added in each well, and the plates were incubated at 37 °C for 24 h. After the incubation period, the developed inhibition zone was measured. The lowest clear zone sizes are scored as heavy metal-resistant strains (Kelany et al., 2019).
Minimum tolerance concentration of bacterial isolates
The highest growth bacterial isolates with different metals were chosen for the determination of the minimum inhibition concentration required for Zn2+, Fe2+, Pb2+, Co2+, Mn2+, Ni2+, and Cd2+ remediation. The resistance was determined by the metal dilution method at a concentration of 0.1 to 35 mM. After the addition of the most potent bacterial isolates in Muller–Hinton agar, the plates were pored and inoculated with different metal concentrations by three replicates, and controls without metals were used. Three-day incubation period at 37 °C was proposed for cultivation. The minimum inhibitory concentration (MIC) is defined as the minimum concentration of the heavy metal solution that prevents the growth of bacterial isolates (Gupta Mahendra et al., 2014).
Identification and characterization of most potent isolates
The most potent isolates were identified biochemically and genetically. The biochemical level was designed by microscopic examination (Ibrahim et al., 2021). The biochemical tests were beta-galactosidase test (ONPG) for lactose fermentation as a tool to differentiate the members of the Enterobacteriaceae, lysine decarboxylase, citrate utilization, hydrogen sulfide production, urease, arginine dihydrolase, tryptophan deaminase, oxidase, ornithine decarboxylase, indole, and Voges–Proskauer. On the other hand, testing different enzyme productions (arabinose, rhamnose, gelatinase, glucose, sorbitol, mannitol, inositol, sucrose, and melibiose) was applied.
A glycerol stock of 20% (glycerol/medium) of pure cultures was prepared and kept for the second identification level, which was genetic identification (Mitra et al., 2018). Identification on gene level was processed. According to the protocol supplied with QIAquick kits (Qiagen, Valencia), genomic DNA and PCR product of 16S rDNA fragment were purified and transferred to the next level. The approach of the Bigdye Terminator V3.1 cycle sequencing kit (PerkinElmer) was applied. The resulting sequence was implemented using the Applied Biosystems3130 genetic analyzer (HITACHI, Japan). Accession numbers for identified strains were given with aid of BLAST® analysis (Basic Local Alignment Search Tool) (Kim et al., 2012). The phylogenetic tree was established by the MegAlign module of LasergeneDNAStar version 12.1 (Abed et al., 2020), and phylogenetic analyses were constructed based on maximum likelihood, neighbor-joining, and maximum parsimony in MEGA6 (Tamura et al., 2013). The identified strains were deposited in the world data center of microbiology (WDCM), Suez Canal University Fungarium (SCUF), Egypt.
Heavy metal assessment after and before bioremediation for water and sediment
Filtration of water samples by a 0.45-m membrane filter was done, and the heavy metals were pre-concentrated and separated from seawater samples by the ammonium pyrrolidine dithiocarbamate (APDC)/methyl isobutyl ketone (MIBK) solvent extraction technique (Eaton et al., 1995; Folk, 1980). Finally, the metals in the organic layer were extracted using 50% HNO3 and collected in a polyethylene bottle to be analyzed by atomic absorption spectrometry (FAAS PerkinElmer model A Analyst 100) for Zn2+, Fe2+, Pb2+, Co2+, Mn2+, Ni2+, and Cd2+. On the other hand, the sediments were dried for 48 h at 60 °C in a thermostatically controlled oven, homogenized with an agate pestle and mortar and sieved using a 63-μm sieve. In a dry Teflon beaker, 0.5 g of fine sediment powder was thoroughly digested at 85 °C with a mixed acid solution containing HNO3:HClO4 (3:1 v/v) according to the method described by Oregioni and Aston (1984). Studied metals were analyzed by FAAS (PerkinElmer model A Analyst 100), and the results were expressed as mg/kg. Each heavy metal was analyzed in three replicates, and the results were presented as mean (Chester et al., 1994; Oregioni & Aston, 1984)
Sieve analysis with carbonate and organic matter determination
Granulometric analysis
To estimate the granulometric analysis; 100 g of each disaggregated day sample was analyzed mechanically by using a standard set of sieves according to Wentworth scale every one phi (Ø) interval. The collected sieve fractions were accurately weighed. The grain size statistical parameters are mean size (MZ), sorting (δI), skewness (SKI), and the kurtosis (KG) according to Folk (1974) and are computed in the BASIC program “GW-BASIC 3.22” (GRSIZE) according to Rząsa and Owczarzak (2015). Varied sized of seven portions were gathered as follows: gravel (Ø-1), very coarse sand (Ø0), coarse sand (Ø1), medium sand (Ø2), fine sand (Ø3), very fine sand (Ø4), and mud (<Ø 4) (Folk, 1980; Rząsa & Owczarzak, 2015).
Geochemical analyses
For the geochemical analyses, about 10g of each sample was ground by agate mortar to less than 80 mesh. Studying the geochemical characteristics of the sediment is designed by measuring total carbonate and total organic matter.
Total carbonate determination
Carbonate matter in terms of CO3% was measured in the target samples. The adjusted weight (1 g) of was thoroughly mixed with 25 ml diluted glacial acetic acid using shaking apparatus overnight. The remained ground samples after incubation were dried, and the difference in weight, before and after incubation, was considered the carbonate content representing as a percentage of the total weight (Dar et al., 2016). The carbonate percentage was calculated upon the next equation:
Total organic matter content
After 2 h of incubation at 550°C, 1 g of each sample was burned to ash. Eventually, the organic matter constituent of each sediment sample was measured from consecutive weight loss (Brenner & Binford, 1988; Liu et al., 2019). Upon the following equation, total organic matter was measured:
Consortium application for drain sewage bioremediation using bacterial isolates
E. kobei, E. cloacae, and E. hormaechei were used for bioremediation of Zn2+, Fe2+, Pb2+, Co2+, Mn2+, Ni2+, and Cd2+ from the water of industrial drainage wastewater by metal concentration 100 %, 200 %, and 300 %. The composition of the medium used was 1000 ml industrial effluent by different concentrations, 1g K2HPO4, 1 g KH2PO4, 0.1 g NaCl2, 1 g NH4NO3, 0.5 g MgSO4·7H20, 10 g yeast extract, 10 g beef extract, and 0.02 g CaCl2. The removal of heavy metals with various concentrations was tested using bacterial isolates, each type separately, and again with three isolates combined for each metal concentration. The prepared flasks were cultivated for 96 h at 37 °C. Bioremediation patterns were measured every 12 h of incubation by absorbance at 600 nm using a Spekol 1900, UV-VIS spectrophotometer, and metal concentration measurement using PerkinElmer A Analyst 100 atomic absorption spectrometer as illustrated in Section 2.6. According to Ijoma et al., 2019, the bacterial isolates were introduced to the MIC test using water from industrial effluent which was replaced by distilled water and added the components of the medium (Ijoma et al., 2019).
Statistical analysis
Standard deviation (±SD) with probability (P<0.05) was calculated for presenting data. The significance of data using the ANOVA test was evaluated by XISTATE (Microsoft, USA) and GraphPad Prism 4 (USA).
Result
Isolation and screening of heavy metal-resistant bacterial isolates
Thirty bacterial isolates were isolated from eight samples of water and sewage that were collected from the main sewage drain in the Al-Adabiya area, Suez, Egypt. All isolated samples were subjected to a growth tolerance test in the presence of different types of heavy metals. After screening, out of these 30 isolates, only 3 bacterial isolates exhibited a varying degree of heavy metal resistance potential against selected heavy metals (Fig. 2a and b). Figure 2 shows the positive and negative resistance of bacterial samples to heavy metals. Table 1 illustrated all isolates’ resistance to the 10 mmol/l concentration of each heavy metal. The most tolerant samples were S4, S5, and S7. Hence, these isolates were selected for further study and identified by PCR sequence analysis.
Figure 1 showed the absorbance and number of bacterial cells in different isolates after 24 and 48 h of incubation. The results showed that samples number four, five, and seven were the growing samples in the presence of heavy metal concentrations. The absorbance of sample number four was 0.3326, 0.9978, and 1.39692 after 0, 24, and 48 h of incubation, and the number of bacteria per ml was 465.64 CFU/ml. Also, the absorbance of sample number five was 0.2976, 0.8928, and 1.24992 after 0, 24, and 48 h of incubation, and the number of bacteria per ml was 416.64 CFU/ml. On the other hand, the absorbance of sample number seven was 0.2944, 0.8832, and 1.23648 after 0, 24, and 48 h of incubation, and the number of bacteria per ml was 412.16 CFU/ml.
Table 1 illustrates the most tolerant bacterial isolates for the presence of 10 mmol of metal ions. Samples four, five, and seven were the most tolerant isolates. Sample four justified the tolerance by zone average of clearance 4, 0, 7, 0, 5, and 0 (mm) for Zn+2, Fe+2, Pb+2, Co+2, Mn+2, Ni+2, and Cd+2 metal, respectively. The lowest tolerance rate was demonstrated for Zn2+, Pb2+, and Mn2+, and the highest was for Fe+2, Co+2, Ni+2, and Cd+2, respectively. Sample five listed tolerance by 5, 0, 0, 6, 0, 4, and 5 (mm) for Zn+2, Fe+2, Pb+2, Co+2, Mn+2, Ni+2, and Cd+2 metal, respectively. The lowest tolerance rate was recorded for Co+2, and the highest was for Cu+2, Fe+2, Pb+2, and Mn+2, respectively. Finally, sample seven recorded tolerance by 0, 0, 5, 4, 0, 5, and 6 (mm) for Zn+2, Fe+2, Pb+2, Co+2, Mn+2, Ni+2, and Cd+2 metal, respectively. The lowest tolerance rate was recorded for Cd+2, and the highest was for Zn+2, Fe+2, and Mn+2 metal, respectively.
Biochemical identification and molecular taxonomy of a selection of heavy metal bioremediation bacterial isolates
Three potent bioremediation isolates (S4, S5, and S7) were extracted and identified using microscopic examination, morphological, biochemically, and biosystems 3130 genetic analyzers. The microscopic examination in supplementary file b figure 2S-b revealed that S4 appeared as a short rod (i), while S5 was a cocci-like structure (ii). S7 showed a typical red shape (iii).
The biochemical tests illustrate differentiation between the three tested potential strains, where ONPG, arginine dihydrolase, lysine decarboxylase, Simmons citrate, tryptophan deaminase, and mannitol played the key elements of differences between those strains; otherwise, all other tests provoked similar results between them. As shown in Table 2, the strain E. cloacae expressed positive signs with lysine decarboxylase, ornithine decarboxylase, citrate utilization, H2S production, urease, tryptophan deaminase, and Voges–Proskauer test, in addition to positive effect for fermentation of glucose, sorbitol, rhamnose, sucrose, and arabinose. On the other side, ONPG, arginine dihydrolase, citrate utilization, H2S, indole, oxidase, gelatinase, mannitol, inositol, and melibiose were negative. E. kobei achieved positive reactions for ONPG, arginine dihydrolase, ornithine decarboxylase, citrate utilization, urease, tryptophan deaminase, Voges–Proskauer test, glucose, mannitol, sorbitol, rhamnose, sucrose, and arabinose with the negative reaction for lysine decarboxylase, H2S, indole, oxidase, gelatinase, inositol, and melibiose. E. hormaechei gave positive reactions for ONPG, arginine dihydrolase, ornithine decarboxylase, citrate utilization, urease, Voges–Proskauer test, glucose, mannitol, sorbitol, rhamnose, sucrose, and arabinose, but the negative reaction was for lysine decarboxylase, H2S, tryptophan deaminase, indole, oxidase, gelatinase, inositol, and melibiose.
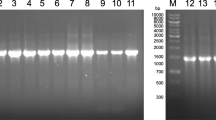
The genetic identification of bacterial isolates was explained using Biosystem 3130 genetic analyzers; this analyzer produced 16S rRNA bases by 1414, 1399, and 1407 for S4, S5, and S7 isolates. The gene bases were identified to genus level (up to 99% identity or better), using available GenBank databases. According to 16S rRNA gene sequence analysis of isolate S4, S5, and S7 compared to Blast which provided the highest homology. The results showed that the isolates under study were similar to Enterobacter spp. and recorded in the NCBI database as E. kobei OM144907 (S4), E. cloacae OM180597 (S5), and E. hormaechei OM181067 (S7) with 99.58, 99.79, and 99.86% similarity percentage. All identified strains are deposited in WDCM with reference numbers SCUF0000311, SCUF0000312, and SCUF0000313 for E. kobei OM144907, E. cloacae OM180597, and E. hormaechei OM181067, respectively. As shown in supplementary file b figure 3S-b-i, the identified strain (E. kobei SCUF0000311) and E. kobei (NZ-JZYH01000051) were in the same clade by 0.72 points with 99.58 % similarity. The most similar strains to our identified strain were Pantoea agglomerans (MW876168), Enterobacter sp. (KU986680), E. ludwigii (MN636653), E. kobei (NZ-LEEC01000015), and P. agglomerans (MW876157). Also, E. cloacae (SCUF0000312) phylogeny is designed in supplementary file b figure 3S-b-ii by similarity 99.79%. Our identified isolates were most similar to Bacterium sp. (MK823507), E. cloacae (KU297784), E. ludwigii (MH001397), Enterobacter sp. (MN540103), P. agglomerans (FJ592995), and Enterobacter sp. (GQ169799). About E. hormaechei (SCUF0000313), it attained 99.86% similarity with Enterobacter sp. (MF401327). E. hormaechei (MW582664), E. hormaechei (MW435507), E. hormaechei (MW582678), Bacterium sp. (MZ045739), E. hormaechei (MT941037), and E. hormaechei (MN428803) which were closely similar to our identified strain (supplementary file b figure 3-b-iii).
Minimum inhibition concentration of tolerant samples
The MICs of the seven metal ions against the studied bacterial isolates were shown in Figure 2. The growth rate of the bacteria exhibited a gradual increase by decreasing metal concentration relative to the control. The concentration of Zn+2, Fe+2, Pb+2, Co+2, Mn+2, Ni+2, and Cd+2 were 0.1, 1, 10, 15, 25, and 35 mmol/l. The MIC for E. kobei and E. cloacae against metals ion were demonstrated by 25 mmol/l for Ni+2, 15 mmol/l for Fe+2 and Mn+2 and 10mmol/l for Zn+2, Pb+2, Co+2, and Cd+2. On the other hand, the MIC for E. hormaechei against metals ion was demonstrated by 15 mmol/l for Ni+2, Fe+2, and Mn+2 and 10 mmol/l for Zn+2, Pb+2, Co+2, and Cd+2. The growth pattern appears to suggest tolerance development or adaptation of bacteria to the presence of heavy metals.
Sediment sieve analysis with carbonate and organic matter
The bioremediation capacity depends on the geochemistry of the drain pathway, which by analysis is described as very coarse silty medium sand with a muddy texture. The percentage of gravel, sand, and mud was 0.00%, 84.4%, and 15.6%, respectively. Grain size statistical parameters such as mean size (Mz), kurtosis (KG), and skewness (SKI) are 2.622, 0.799, and 0.165. The total organic matter % and total carbonate % of the drain were 31.5 and 20.54 % (Table 3). On the other hand, the analysis of marine sediment achieved poorly sorted very coarse sand by gravel, sand, and mud 0.00, 98.6, and 1.4 with mean size (Mz), kurtosis (KG), and skewness (SKI) 1.099, 0.21 and 0.71, respectively.
The metal concentrations of drain and marine sediment are different considering the sources of the pollutants. The concentration of Fe+2, Mn+2, Zn+2, Pb+2, Cd+2, Ni+2, and Co+2 metals were 2.71, 5.84, 1.68, 92.06, 3.80, 72.06, and 12.48 mg/g; nevertheless, in marine sediment, the Fe+2, Mn+2, Zn+2, Pb+2, Cd+2, Ni+2, and Co+2 concentrations were 5.6, 5.3, 29.5, 5.9, 1.64, 24.1, 2.63, and 1.6 μg/g, respectively (Table 3).
Application of bacterial strains for bioremediation of drain sewage water
The consortium test for metal removal at 100%, 200%, and 300% concentrations is expressed by absorbance measuring at 600 nm and metal concentration measuring using atomic absorption every 12 h until 96 h of incubation. The results are represented in Figures 3, 4, 5, 6, 7 , 8 and 9 for Zn+2, Pb+2, Ni+2, Mn+2, Fe+2, Co+2, and Cd+2, respectively.
As illustrated in Figure 3, starting with no remediation, where the concentration before any treatment was 54.2 μg/l, approximately a complete removal of Zn+2 with a removal percentage of 99% was achieved after 96-h incubation period for the examined strains and their consortium as well. However, E. cloacae removed about 65% of the doubling load of Zn+2 in the polluted sample after an incubation period of 96 h. In addition, it removed about 47% of Zn+2 from the tripling concentration of the sample after 84-h incubation time within the stationary growth phase in all cases.
Having an initial concentration of 15.12 μg/l of Pb+2, all tested potential strains exhibited a high efficiency of Pb+2 removal (99%) after 96-h incubation time during a stationary growth phase, even with their consortium. But, E. cloacae was the one that succeeded in removing about 75% of Pb+2 from doubling the concentration of Pb+2 after 96-h incubation period, while E. kobei was the one that removed about 51% of the tripling load of Pb+2 concentration in the polluted sample after 96 h within the stationary growth phase (Figure 4).
In Figure 5, about 93% removal of Ni+2 from the initial concentration load 3.32 μg/l was reported after incubation of the polluted sample with the potential strains and their consortium as well for 96-h incubation period. Yet, E. cloacae alone showed a great potential to remove Ni+2 from doubling and tripling concentration of Ni+2 by 66% and 46% after 96 and 84 h, respectively, within its stationary growth phase.
In Figure 6, starting with a concentration of 0.484 μg/l of Mn+2 as the initial loading sample, E. cloacae alone showed a remarkable efficiency to remove Mn+2 (91%) after 96-h incubation time during the stationary growth phase, and such efficiency was kept steady with doubling and tripling load of Mn+2 concentration in the sample with removal percentage 57% and 48%, respectively, after 84-h incubation time during the stationary growth phase.
Polluted sample with Fe+2 having a concentration of 44.76 μg/l was bio-remediated to approximately 97% removal with equal efficiency for all tested strains and their consortium after 96-h incubation time with the stationary growth phase of them. Nevertheless, 63% removal of doubling the Fe+2 concentration after 96-h incubation time was reported by E. hormaechei. Yet, E. cloacae were the supreme of removing Fe+2 in all incubation periods except 96-h incubation measurement. On the other side, equal removal efficiency (47%) of tripling the Fe+2 concentration from the polluted sample was done by the three tested potential strains in addition to their consortium during the stationary phase of their growth (Figure 7).
In Figure 8, the highest removal of Co+2 (84%), where the initial loading concentration (100%) was 0.765 μg/l, was achieved by E. cloacae after 96-h incubation period; however, E. kobei removed about 83%, as well, of polluted sample from Co+2 after 84 h, doubling the concentration of Co+2; E. hormaechei succeeded to remove about 63% of Co+2 after 96 h, while E. cloacae removed about 57% after 84 h. with increasing the concentration of pollutant representing as Co+2 to triple the concentration in the original polluted sample; E. cloacae removed about 46% of Co+2 after 48 h. All successive removal was determined during the stationary phase of all tested microbial growth.
In Figure 9, the highest removal percentage of Cd+2 starting from loading concentration 1.142 μg/l was achieved between 94 and 96%) from a polluted sample using the three potential strains separately and their consortium as well after 96-h incubation period, where the stationary phase of their growth has occurred. However, with doubling the Cd+2 concentration in the polluted sample, E. cloacae expressed the highest efficiency of removal percentage (70%) after 96-h incubation time within the stationary growth phase. With more loading of Cd+2 concentration in the treated sample reaching tripling the original concentration, E. cloacae removed about 48% of Cd+2 after 48-h incubation time within the stationary growth phase. The data of solo and consortium species removal is illustrated in the supplementary file table 1S-a, 2S-a, 3S-a, and 4S-a.
The microbial bioremediation of Zn+2, Pb+2, Ni+2, Mn+2, Fe+2, Co+2, and Cd+2 using (i) E. kobei Wdcm scuf0000311, (ii) E. cloacae Wdcm scuf0000312, (iii) E. hormaechei Wdcm scuf0000312, and (iv) consortium culture of previously applied strains on different concentrations (a) polluted sample representing 100%; (b) doubled heavy metal concentration in the same sample representing 200%, of the original polluted sample; and (c) tripled concentration of heavy metal concentration representing 300%, of the original polluted sample. The standard deviation was calculated for each record as SD 0.05.
Discussion
The more industrial activities discharged without any treatment, the more pollution and toxic effects on the relevant surrounding environment get. This would be the major reason for spreading the pollution. Time-consuming and charging a high cost to mechanically remove the heavy metal contaminants result in the deviation of scientists’ thoughts towards a practical solution that focuses on using bacterial cells possessing multiple mechanisms for heavy metal removal. The current study succeeded in isolating and purifying three bacterial isolates genetically identified as E. kobei (SCUF0000311), E. cloacae (SCUF0000312), and E. hormaechei (SCUF0000313) and having a potential resistance to high concentrations of Zn2+, Fe2+, Pb2+, Co2+, Mn2+, Ni2+, and Cd2+ metals using accumulation property. Lately, Banerjee et al. (2015) have reported E. cloacae as a potent strain to accumulate lead, cadmium, and nickel, whereas Bestawy et al. (2013) have removed different heavy metals from contaminated domestic–industrial effluent with aid of eight resistant indigenous bacteria isolated from activated sludge as well as Rahman et al. (2015) who have reported the possibility of chromium removal from local human activities (industries, agriculture, forest farming, mining, and metallurgy) using E. cloacae B2-D HA. These manuscripts has been studied and identified to have various resistance mechanisms including transport channels and compartmentalization within the cell (Banerjee et al., 2015; Bestawy et al., 2013; Rahman et al., 2015).
Varied heavy metal removal mechanisms have been reported such as bacterial cell wall attachment, siderophores production for chelation, and heavy metal metabolic transportation (Ahemad, 2012; Schalk et al., 2011). As reported in the current study, the minimum inhibitory concentration of E. kobei (SCUF0000311), E. cloacae (SCUF0000312), and E. hormaechei (SCUF0000313) against Ni2+, Fe2+, and Mn2+ was recorded to be 15mmol/l compared to Zn2+, Pb2+, Co2+, and Cd2+ with 10mmol/l. Previous studies have reported MIC of Bacillus carotarum, B. cereus, B. lentus, and B. licheniformis isolated from Jabalpur, India, against lead, zinc, and chromium by 1% and 0.01% (Gupta Mahendra et al., 2014). Moreover, E. cloacae B2-DHA has recorded MIC value against chromium as 1000 μg/mL−1 (Rahman et al., 2015).
Our study encompasses a vast amount of information about the bioremediation process of a considerable number of heavy metals. This study approached the measurement of bioremediation in an innovative way by experimenting with the removal of heavy elements separately by E. kobei (SCUF0000311) and E. cloacae (SCUF0000312) and E. hormaechei (SCUF0000313) and by combining the three strains into one sample and testing them individually. This method had not been previously addressed by any of the previous scientists, as we have in our current study, resulting in a precise analysis of heavy element removal percentages using the mentioned strains. Poornima et al. (2014) and Pandey et al. (2011) achieved a similar concept in our study without our sequence work by isolating E. coli PS01 and Bacillus sp., both of which can withstand high concentrations of chromium, lead, and arsenic (Pandey et al., 2011; Poornima et al., 2014). In the study conducted by Rani et al. (2010), three bacterial isolates, namely, Bacillus sp., Pseudomonas sp., and Micrococcus sp., were isolated, and their bioaccumulation capacities were reported as follows: 69.34% for copper, 90.41% for cadmium, and 84.27% for lead. Similarly, Ahemad and Malik (2011) documented the accumulation of various metals such as lead, chromium, mercury, and zinc by multiple bacterial species isolated from agricultural fields and wastewater. In contrast, our study revealed that the bacterial strain E. cloacae B1 exhibited significantly higher lead accumulation capacity compared to cadmium and nickel.
As previously documented by numerous researchers, various bacterial strains have been shown to possess metal-reducing capabilities, demonstrating their potential for biotransformation and the ability to reduce varying amounts of chromium in the medium. Thacker et al. (2007) reported the existence of a Gram-negative strain of Brucella sp. with the capacity to reduce chromium levels in contaminated sources. This strain’s resistance to high concentrations of metals and its proficiency in reducing this toxic metal make it a promising candidate for bioremediation purposes. Additionally, scientists can identified and characterized three highly efficient metal-reducing bacterial strains, namely Bacillus cereus, Bacillus fusiformis, and Bacillus sphaericus, which were isolated from metal-polluted landfills and evaluated for in vitro metal reduction (Desai et al., 2008; Zhang & Wang, 2021). This aligns with what we have reached through our current study, which allows us to assert the potential use of microbes for the removal of heavy elements from industrial wastewater.
Metal concentrations of Fe+2, Mn+2, Zn+2, Pb+2, Cd+2, Ni+2, and Co+2 were 2.71, 5.84, 1.68, 92.06, 3.80, 72.06, and 12.48 μg/g in a sediment layer, respectively. Maslennikova et al. (2012) have indicated that within the smaller grain size where the higher surface area exists, the more heavy metal content to be there. Also, previous studies have revealed that organic matter hydrolysis in bottom sediments could be another source for adsorbing heavy metals on sediment grains that would be later liberated into the surrounding environment via desorption, microbial activities, substitution, or dissolution due to any alter of pH levels or redox potential processes, which in turn would reflect on water quality and surrounding aquatic ecosystem (Maslennikova et al., 2012; Yang et al., 2020; Zamani Hargalani et al., 2014).
The nature of the drain sediment was different from marine, which explains the accumulation of pollutants in the drain sediment leading to the appearance of soil as clay and muddy allowing for heavy metal accumulation. Dixit et al. (2015) reported that a heavily polluted soil allows water droplets to adhesion to the hydrophobic layer, and this prevents the wetting of the soil aggregates (Dixit et al., 2015).
During this study, the bacterial strains that were isolated in this study area could not reduce the field metal percentage. By the availability of suitable conditions for bacterial growth, isolated strains were adapted for metal high percentages in the presence of growth factors and nutrition. It is noteworthy that the nature of the clay soil in the drain area does not allow aerobic bacterial growth but allows anaerobic bacteria enumeration (Chen et al., 2021). Wellsbury et al. (2002) recognized that small pores restrict bacteria movement and activity, limit nutrient transport, diminish space availability, slow the rate of division, and lead to reduced biodiversity. So, the most species of bacteria isolated in this study were Enterobacter sp. (Chen et al., 2021; Wellsbury et al., 2002).
It was observed that toxic sediments including decaying organic matters play a vital role in controlling the binding of existing heavy metals to sediment grains as well as the bioavailability of heavy metals with different toxicity and safety levels. However, quantitative measurement of organic matter content is rarely analyzed in contaminant studies. On the other side, it was found that the composition of organic matter varies widely within the available organic matter content offering diverse effects (Baran & Tarnawski, 2015; Chiriluș et al., 2022).
The concept of microbial heavy metal bioremediation has been evaluated via biosorption, bioaccumulation, bioprecipitation, or biomineralization. Those are the milestones of any microbial remediation so far, and the metabolic pathway of each differs from microbial strain to another (Lin & Lin, 2005; Sreedevi et al., 2022). The current study has revealed that, upon studied strains, Enterobacter spp. include potent strains for heavy metal bioremediation. Out of three examined Enterobacter strains (E. kobei SCUF0000311, E. cloacae SCUF0000312, and E. hormaechei SCUF0000313), E. cloacae (SCUF0000312) proved to be the one with high capability to bioremediate a broad spectrum of heavy metals including the current study with the privilege to bioremediate high concentrations as doubling and tripling the original waste concentration with efficient time factor in comparison with other previous studies of Enterobacter spp. This study showed that MIC for E. kobei and E. cloacae against (Ni+2), (Mn+2, Fe+2) and (Zn+2, Pb+2, Co+2, Cd+2) were 25, 15, and 10 mmol/l, respectively, while MIC for E. hormaechei against (Mn+2, Ni+2, Fe+2) and (Zn+2, Pb+2, Co+2, Cd+2) were 15 and 10 mmol/l.
Enterobacter species have been registered by Fadzli et al. (2021) as a potent species for heavy metal remediation recording high removal efficiency of Pb+2, Cd+2, and Cr+3 as 90.14, 88.00, and 90.34%, respectively, within 30-day incubation (Fadzli et al., 2021).
E. cloacae have been observed as an efficient microbial biosorbent giving a high uptake concentration of Pb+2 (2.3 mmoles) from the initial concentration (7.2 mmol) (Kang et al., 2015). In addition, E. cloacae have been found to have MIC (1000 ug/ml) with Cr+2 having a mechanism of intracellular accumulation of heavy metal and recording 81% of Cr+2 reduction from the liquid medium after 120-h incubation period (Rahman et al., 2015). Banerjee et al. (2015) have reported that the MIC of E. cloacae towards Pb+2, Cd+2, and Ni+2 was 1100, 900, and 700 ppm, respectively. Consequently, the high efficiency of bioaccumulation in percentage with those heavy metals has been recorded as Pb+2 (95.25%), Cd+2 (64.17%), and Ni+2 (36.77%) (Banerjee et al., 2015). Moreover, Abdollahi et al. (2020) have reported that E. cloacae had MIC 3000 ug/ml and 50 ug/ml against Pb+2 and Cd+2 with accumulation capacity 45ug Pb+2/ml and 30ug Cd+2/ml. Also, Ghosh et al. (2022) have reported that E. cloacae expressed a high potency of tolerance towards high concentrations of Cd+2 (4000 μg/ml), Pb+2 (3312 μg/ml), and As+3(1500 μg/ml), where the removal efficiency of Cd+2 was recorded 72.11% (Abdollahi et al., 2020; Ghosh et al., 2022).
With a few reports on the capability of E. hormaechei (SCUF0000313) and E. kobei (SCUF0000311) to bioremediate heavy metals, Heidari et al. (2020) have found that E. hormaechei exposed a high efficiency of uptake towards Ni+2 than Pb+2 and Cd+2. Abdollahi et al. (2020) found that E. kobei had MIC 3000 μg/ml and 50 μg/ml towards Pb+2 and Cd+2, respectively, in addition to an accumulation capacity of 25 μg Pb+2/ml and 20μg Cd+2/ml (Abdollahi et al., 2020; Heidari et al., 2020).
Overall, among the tested potential Enterobacter spp. for heavy metal remediation, E. cloacae (SCUF0000312) has proved to be the most potent strain for water treatment in a sufficient way.
Conclusion
In conclusion, the study presented here highlights the critical role that bacterial strains, particularly Enterobacter spp., can play in the bioremediation of heavy metals from polluted environments. The traditional methods for removing heavy metal contaminants are often time-consuming and costly. The research conducted in this study isolated and identified three Enterobacter strains, namely, E. kobei (SCUF0000311), E. cloacae (SCUF0000312), and E. hormaechei (SCUF0000313), which exhibited high resistance to a range of heavy metals, including zinc, lead, cobalt, cadmium, and others. Of these strains, E. cloacae (SCUF0000312) emerged as particularly effective in bioremediation efforts, surpassing other Enterobacter species in terms of both efficiency and capacity. Different heavy metal removal mechanisms have been reported, including bacterial cell wall attachment, siderophores production for chelation, and heavy metal metabolic transportation. Furthermore, this study introduced an innovative approach to assessing heavy metal removal by experimenting with individual strains and their combined effectiveness. This method allowed for a precise analysis of heavy metal removal percentages using these specific bacterial strains, which had not been previously explored in such detail. The study area is characterized by its clay and muddy composition, which presented challenges for aerobic bacterial growth. However, anaerobic bacterial enumeration was possible, underscoring the importance of environmental factors in shaping bacterial activity and metal removal capabilities. The findings from this study contribute to the growing body of research on microbial bioremediation and emphasize the potential of Enterobacter spp., particularly E. cloacae (SCUF0000312), as valuable tools in addressing heavy metal pollution in industrial wastewater. The versatility and efficiency demonstrated by these bacterial strains offer promising avenues for the development of sustainable and cost-effective solutions to mitigate the harmful effects of heavy metal contamination on the environment. Continued research in this field can lead to more effective bioremediation strategies that help protect ecosystems and human health.
Data availability
The raw data supporting the conclusions of this manuscript would be available by the authors, without undue reservation, to any qualified researcher.
Abbreviations
- Zn2+ :
-
zinc
- Fe2+ :
-
iron
- Pb2+ :
-
lead
- Co2+ :
-
cobalt
- Mn2+ :
-
manganese
- Ni2+ :
-
nickel
- Cd2+ :
-
cadmium
- DNA:
-
deoxyribonucleic acid
- E. kobei :
-
Enterobacter kobei
- E. cloacae :
-
Enterobacter cloacae
- E. Hormaechei :
-
Enterobacter hormaechei
- K2HPO4 :
-
dipotassium hydrogen orthophosphate
- KH2PO4 :
-
potassium dihydrogen orthophosphate
- NaCl:
-
sodium chloride
- NH4NO3 :
-
ammonium nitrate
- MgSO4 :
-
magnesium sulfate
- Pb(CH3COO)4 :
-
lead tetraacetate
- CuSO4 :
-
copper sulfate
- ZnSO4 :
-
zinc sulfate
- Co(NO3)2·6 H2O:
-
cobalt nitrate anhydrase
- CaCl2 :
-
calcium chloride
- H2O:
-
water
- h:
-
hours
- °C:
-
degree Celsius
- rpm:
-
rotation per minute
- OD:
-
optical density
- CFU/mL:
-
colony-forming unit per mill
- μl:
-
micron
- mM:
-
mill mole
- MIC:
-
minimum inhibition concentration
- ONBG:
-
O-nitrophenyl-beta-D-galactopyranoside
- rDNA:
-
ribosomal deoxyribonucleic acid
- APDC:
-
ammonium pyrrolidine dithiocarbamate
- MIBK:
-
methyl isobutyl ketone
- HNO3 :
-
nitric acid
- HClO4 :
-
perchloric acid
- FAAS:
-
flame atomic absorption spectroscopy
- Ø:
-
one phi
- MZ:
-
mean size
- δ:
-
sorting
- SKI:
-
skewness
- KG:
-
kurtosis
- CO3:
-
carbonate
- TOM:
-
total organic matter
References
Abdollahi, S., Golchin, A., & Shahryari, F. (2020). Lead and cadmium-resistant bacterial species isolated from heavy metal-contaminated soils show plant growth-promoting traits. International Microbiology, 23, 625–640.
Abed, A. H., El-Seedy, F. R., Hassan, H. M., Nabih, A. M., Khalifa, E., Salem, S. E., Wareth, G., & Menshawy, A. M. (2020). Serotyping, genotyping and virulence genes characterization of Pasteurella multocida and Mannheimia haemolytica isolates recovered from pneumonic cattle calves in North Upper Egypt. Veterinary Sciences, 7, 174.
Ahemad, M. (2012). Implications of bacterial resistance against heavy metals in bioremediation: A review. Journal of Institute of Integrative Omics and Applied Biotechnology(IIOAB), 3, 3.
Ahemad, M., & Malik, A. (2011). Bioaccumulation of heavy metals by zinc resistant bacteria isolated from agricultural soils irrigated with wastewater. Bacteriology Journal, 2, 12–21.
Alloway, B. J. (2012). Heavy metals in soils: Trace metals and metalloids in soils and their bioavailability (Vol. 22). Springer Science & Business Media.
Ayaz, T., Khan, S., Khan, A. Z., Lei, M., & Alam, M. (2020). Remediation of industrial wastewater using four hydrophyte species: A comparison of individual (pot experiments) and mix plants (constructed wetland). Journal of Environmental Management, 255, 109833.
Banerjee, G., Pandey, S., Ray, A. K., & Kumar, R. (2015). Bioremediation of heavy metals by a novel bacterial strain Enterobacter cloacae and its antioxidant enzyme activity, flocculant production, and protein expression in presence of lead, cadmium, and nickel. Water, Air, & Soil Pollution, 226, 1–9.
Baran, A., & Tarnawski, M. (2015). Assessment of heavy metals mobility and toxicity in contaminated sediments by sequential extraction and a battery of bioassays. Ecotoxicology, 24, 1279–1293.
Bestawy, E. E., Helmy, S., Hussien, H., Fahmy, M., & Amer, R. (2013). Bioremediation of heavy metal-contaminated effluent using optimized activated sludge bacteria. Applied Water Science, 3, 181–192.
Brenner, M., & Binford, M. W. (1988). Relationships between concentrations of sedimentary variables and trophic state in Florida lakes. Canadian Journal of Fisheries and Aquatic Sciences, 45, 294–300.
Chen, M., Li, Y., Jiang, X., Zhao, D., Liu, X., Zhou, J., He, Z., Zheng, C., & Pan, X. (2021). Study on soil physical structure after the bioremediation of Pb pollution using microbial-induced carbonate precipitation methodology. Journal of Hazardous Materials, 411, 125103.
Chester, R., Lin, F.-J., & Basaham, A. (1994). Trace metal solid state speciation changes associated with the down-column fluxes of oceanic particulates. Journal of the Geological Society, 151, 351–360.
Chiriluș, G. V., Lakatos, E. S., Bălc, R., Bădărău, A. S., Cioca, L. I., David, G. M., & Roşian, G. (2022). Assessment of organic carbon sequestration from Romanian degraded soils: Livada Forest Plantation Case Study. Atmosphere, 13, 1452.
Dar, M. A., El-Metwally, M. E., & El-Moselhy, K. M. (2016). Distribution patterns of mobile heavy metals in the inshore sediments of the Red Sea. Arabian Journal of Geosciences, 9, 1–14.
Desai, C., Jain, K., & Madamwar, D. (2008). Evaluation of in vitro Cr (VI) reduction potential in cytosolic extracts of three indigenous Bacillus sp. isolated from Cr (VI) polluted industrial landfill. Bioresource Technology, 99, 6059–6069.
Dixit, R., Malaviya, D., Pandiyan, K., Singh, U. B., Sahu, A., Shukla, R., Singh, B. P., Rai, J. P., Sharma, P. K., & Lade, H. (2015). Bioremediation of heavy metals from soil and aquatic environment: An overview of principles and criteria of fundamental processes. Sustainability, 7, 2189–2212.
Eaton, A.D., Clesceri, L.S., Greenberg, A., Franson, M., (1995). APHA. AWWA, WEF, Standard methods for the examination of water and wastewater. 19th edn, Washington, DC, USA.
Espinosa-Ortiz, E. J., Rene, E. R., Pakshirajan, K., van Hullebusch, E. D., & Lens, P. N. (2016). Fungal pelleted reactors in wastewater treatment: Applications and perspectives. Chemical Engineering Journal, 283, 553–571.
Fadzli, F. S., Rashid, M., Yaqoob, A. A., & Ibrahim, M. N. M. (2021). Electricity generation and heavy metal remediation by utilizing yam (Dioscorea alata) waste in benthic microbial fuel cells (BMFCs). Biochemical Engineering Journal, 172, 108067.
Figueira, E. M., Gusmão Lima, A. I., & Pereira, S. I. A. (2005). Cadmium tolerance plasticity in Rhizobium leguminosarum bv. viciae: Glutathione as a detoxifying agent. Canadian Journal of Microbiology, 51, 7–14.
Folk, R. L. (1980). Petrology of sedimentary rocks. Hemphill publishing company.
Ghosh, A., Pramanik, K., Bhattacharya, S., Mondal, S., Ghosh, S. K., & Maiti, T. K. (2022). A potent cadmium bioaccumulating Enterobacter cloacae strain displays phytobeneficial property in Cd-exposed rice seedlings. Current Research in Microbial Sciences, 3, 100101.
Guagliardi, I., Apollaro, C., Scarciglia, F., & De Rosa, R. (2013). Influence of particle-size on geochemical distribution of stream sediments in the Lese river catchment, southern Italy. BASE.
Gupta Mahendra, K., Kiran, K., Amita, S., & Shikha, G. (2014). Bioremediation of heavy metal polluted environment using resistant bacteria. Journal of Environmental Research And Development, 8, 883–889.
Heidari, P., Sanaeizade, S., & Mazloomi, F. (2020). Removal of nickel, copper, lead and cadmium by new strains of Sphingomonas melonis e8 and Enterobacter hormaechei ww28. Journal of Applied Biotechnology Reports, 7, 208–214.
Ibrahim, A. M., Hamouda, R. A., El-Naggar, N. E.-A., & Al-Shakankery, F. M. (2021). Bioprocess development for enhanced endoglucanase production by newly isolated bacteria, purification, characterization and in-vitro efficacy as anti-biofilm of Pseudomonas aeruginosa. Scientific Reports, 11, 1–24.
Ijoma, G. N., Selvarajan, R., Oyourou, J.-N., Sibanda, T., Matambo, T., Monanga, A., & Mkansi, K. (2019). Exploring the application of biostimulation strategy for bacteria in the bioremediation of industrial effluent. Annals of Microbiology, 69, 541–551.
Ilavský, J., Barloková, D., & Munka, K. (2015). Antimony removal from water by adsorption to iron-based sorption materials. Water, Air, & Soil Pollution, 226, 1–8.
Kang, C.-H., Oh, S. J., Shin, Y., Han, S.-H., Nam, I.-H., & So, J.-S. (2015). Bioremediation of lead by ureolytic bacteria isolated from soil at abandoned metal mines in South Korea. Ecological Engineering, 74, 402–407.
Kelany, M. S., Beltagy, E. A., Khalil, M. A. E.-F., El-Shenawy, M. A., & El-Shouny, W. A. E.-F. (2019). Isolation, characterization, and detection of antibacterial activity of a bioactive compound produced by marine Bacillus sp. MH20 from Suez Bay, Egypt. Novel Research in Microbiology Journal, 3, 258–270.
Kim, O.-S., Cho, Y.-J., Lee, K., Yoon, S.-H., Kim, M., Na, H., Park, S.-C., Jeon, Y. S., Lee, J.-H., & Yi, H. (2012). Introducing EzTaxon-e: A prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. International journal of systematic and evolutionary microbiology, 62, 716–721.
Lin, C.-C., & Lin, H.-L. (2005). Remediation of soil contaminated with the heavy metal (Cd2+). Journal of Hazardous Materials, 122, 7–15.
Liu, B., Schieber, J., Mastalerz, M., & Teng, J. (2019). Organic matter content and type variation in the sequence stratigraphic context of the Upper Devonian New Albany Shale, Illinois Basin. Sedimentary Geology, 383, 101–120.
Maslennikova, S., Larina, N., & Larin, S. (2012). The effect of sediment grain size on heavy metal content. Lakes Reservoirs and Ponds, 6, 43–54.
Mitra, S., Pramanik, K., Sarkar, A., Ghosh, P. K., Soren, T., & Maiti, T. K. (2018). Bioaccumulation of cadmium by Enterobacter sp. and enhancement of rice seedling growth under cadmium stress. Ecotoxicology and Environmental Safety, 156, 183–196.
Muñoz, A., Ruiz, E., Abriouel, H., Gálvez, A., Ezzouhri, L., Lairini, K., & Espínola, F. (2012). Heavy metal tolerance of microorganisms isolated from wastewaters: Identification and evaluation of its potential for biosorption. Chemical Engineering Journal, 210, 325–332.
Mustapha, M. U., & Halimoon, N. (2015). Microorganisms and biosorption of heavy metals in the environment: A review paper. Journal of Microbial & Biochemical Technology, 7, 253–256.
Oregioni, B., & Aston, S. (1984). The determination of selected trace metals in marine sediments by flameless/flame atomic absorption spectrophotometry. IAEA Manaco Laboratory. Water Resources Management, II, 11.
Pandey, S., Saha, P., Biswas, S., & Maiti, T. K. (2011). Characterization of two metal resistant Bacillus strains isolated from slag disposal site at Burnpur, India. Journal of Environmental Biology, 32, 773.
Paul, A., & Mukherjee, S. K. (2016). Enterobacter asburiae KUNi5, a nickel resistant bacterium for possible bioremediation of nickel contaminated sites. Polish Journal of Microbiology, 65.
Poornima, M., Kumar, R. S., & Thomas, P. D. (2014). Isolation and molecular characterization of bacterial strains from tannery effluent and reduction of chromium. International Journal of Current Microbiology and Applied Sciences, 3, 530–538.
Rahman, A., Nahar, N., Nawani, N. N., Jass, J., Hossain, K., Saud, Z. A., Saha, A. K., Ghosh, S., Olsson, B., & Mandal, A. (2015). Bioremediation of hexavalent chromium (VI) by a soil-borne bacterium, Enterobacter cloacae B2-DHA. Journal of Environmental Science and Health, Part A, 50, 1136–1147.
Rahman, Z., Thomas, L., & Singh, V. P. (2019). Biosorption of heavy metals by a lead (Pb) resistant bacterium, Staphylococcus hominis strain AMB-2. Journal of Basic Microbiology, 59, 477–486.
Rani, M. J., Hemambika, B., Hemapriya, J., & Kannan, V. R. (2010). Comparative assessment of heavy metal removal by immobilized and dead bacterial cells: A biosorption approach. African Journal of Environmental Science and Technology, 4.
Rząsa, S., & Owczarzak, W. (2015). Methods for the granulometric analysis of soil for science and practice. Polish Journal of Soil Science, 46, 1.
Schalk, I. J., Hannauer, M., & Braud, A. (2011). New roles for bacterial siderophores in metal transport and tolerance. Environmental Microbiology, 13, 2844–2854.
Sreedevi, P., Suresh, K., & Jiang, G. (2022). Bacterial bioremediation of heavy metals in wastewater: A review of processes and applications. Journal of Water Process Engineering, 48, 102884.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., & Kumar, S. (2013). MEGA6: Molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 30, 2725–2729.
Thacker, U., Parikh, R., Shouche, Y., & Madamwar, D. (2007). Reduction of chromate by cell-free extract of Brucella sp. isolated from Cr (VI) contaminated sites. Bioresource Technology, 98, 1541–1547.
Verma, S., & Kuila, A. (2019). Bioremediation of heavy metals by microbial process. Environmental Technology & Innovation, 14, 100369.
Wellsbury, P., Mather, I., & Parkes, R. J. (2002). Geomicrobiology of deep, low organic carbon sediments in the Woodlark Basin, Pacific Ocean. FEMS Microbiology Ecology, 42, 59–70.
Yang, H. J., Jeong, H. J., Bong, K. M., Kang, T.-W., Ryu, H.-S., Han, J. H., Yang, W. J., Jung, H., Hwang, S. H., & Na, E. H. (2020). Organic matter and heavy metal in river sediments of southwestern coastal Korea: Spatial distributions, pollution, and ecological risk assessment. Marine Pollution Bulletin, 159, 111466.
Zamani Hargalani, F., Karbassi, A., Monavari, S. M., & Abroomand Azar, P. (2014). A novel pollution index based on the bioavailability of elements: A study on Anzali wetland bed sediments. Environmental Monitoring and Assessment, 186, 2329–2348.
Zhang, S., & Wang, J. (2021). Removal of chlortetracycline from water by immobilized Bacillus subtilis on honeysuckle residue–derived biochar. Water, Air, & Soil Pollution, 232, 236.
Acknowledgements
The authors are grateful to the National Institute of Oceanography and Fisheries Science, Egypt, for providing the space and possibilities for this research.
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB). The National Institute of Oceanography and Fisheries, Egypt, financially supported the research.
Author information
Authors and Affiliations
Contributions
K.M., E.M., E.A., and B.E. made significant contributions to the design of the practical parts and the writing of the manuscript. K.M., E.M., and E.A. provided water and sediment sample collection. K.M. performed the bacterial isolation, identification, screening for bioremediation, and MIC test and drafted the manuscript. E.M. performed metal analysis after and before bioremediation. E.A. performed the geological analysis of sediment samples. B.E. designed and implemented the solo and consortium tests for bioremediation and interpreted the data. All authors read, reviewed, and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable. All authors have read, understood, and have complied as applicable with the statement on “Ethical responsibilities of Authors” as found in the Instructions for Authors.
Consent for publication
Not applicable
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kelany, M.S., El-sawy, M.A., El-Gendy, A.R. et al. Bioremediation of industrial wastewater heavy metals using solo and consortium Enterobacter spp.. Environ Monit Assess 195, 1357 (2023). https://doi.org/10.1007/s10661-023-11951-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10661-023-11951-x