Abstract
Background
Bromodomain-containing protein 4 (BRD4) is a reader of histone acetylation and is associated with a variety of diseases.
Aim
To investigate the expression level of BRD4 in esophageal squamous cell carcinoma (ESCC), its prognostic value and its relationship with immune infiltration.
Methods
The study included 94 ESCC patients from The Cancer Genome Atlas (TCGA) database and 179 ESCC patients from Affiliated Hospital 2 of Nantong University. The expression levels of proteins in tissue microarray were detected by immunohistochemistry. The prognostic factors were analyzed by Kaplan–Meier curve and univariate and multivariate cox regression. The ESTIMATE website was used to calculate the stromal, immune and ESTIMATE score. CIBERSORT was used to calculate the abundance of immune infiltrates. Spearman and Phi coefficient were used for correlation analysis. The TIDE algorithm was used to predict treatment response to immune checkpoint blockade.
Results
BRD4 is up-regulated in ESCC, and high BRD4 expression level is associated with poor prognosis and adverse clinicopathological features. In addition, the monocyte count, systemic inflammatory-immunologic index, platelet-lymphocyte ratio, and monocyte-lymphocyte ratio in the BRD4 high expression level group were higher than in the low expression level group. Finally, we found that BRD4 expression level correlated with immune infiltration and that it was inversely correlated with infiltration of CD8 + T cells. Higher TIDE scores in the BRD4 high expression group than in the low expression group.
Conclusion
BRD4 is associated with poor prognosis and immune infiltration in ESCC, and may be a potential biomarker for prognosis and immunotherapy application.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Esophageal cancer is the sixth leading cause of cancer death in the world whose main histological types include esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma [1]. ESCC is regarded as the major type of esophageal cancer in China, accounting for about 90% of all types [2]. Although the treatment methods of ESCC, including surgical resection combined with radiotherapy and chemotherapy, molecular targeted therapy and immunotherapy have been continuously improved, it can be seen that the prognosis of some patients is still poor [3]. Therefore, finding new biomarkers, stratifying risks of patients, and timely and accurate adjuvant treatment of high-risk patients are expected to effectively improve the prognosis.
Epigenetic alterations promote tumor occurrence and development by affecting multiple carcinogenic and tumor suppressor gene pathways and by activating, differentiating, and functioning of immune cells [4]. Bromodomain-containing protein 4 (BRD4) is an important member of the bromodomain and extra-terminal domain (BET) family proteins, an epigenetic reader linked to histone acetylation and synchronized with gene transcription [5]. A large number of studies have shown that BRD4 is related to the occurrence and development of various tumors. According to reports by Qin et al., BRD4 regulates snail-driven metastasis of gastric cancer cells by preventing FBXL14 or β-Trcp1 from binding to snail ubiquitination and stabilizing snail protein in an acetylation-dependent manner [6]. In addition, BRD4 also regulates the transcription of a variety of pro-inflammatory and immunomodulatory genes, and has become an significant therapeutic target for diseases related to immune system and inflammation [7]. Previous studies have proved that BRD4 can be used as a potential biomarker to evaluate the prognosis of breast cancer, liver cancer, bladder cancer and other tumors [8,9,10]. It has demonstrated in the latest published literature that BRD4 promotes the growth of ESCC. Thus, the inhibition of BRD4 can inhibit the growth of tumor cells [11, 12]. However, the prognostic value of BRD4 in ESCC and its role in immune microenvironment still remain unclear.
This study evaluated the prognostic value in patients with ESCC by detecting the expression level of BRD4 in patients with ESCC through The Cancer Genome Atlas (TCGA) database and immunohistochemical methods. The correlation between BRD4 and immune infiltration in ESCC was also further discussed in TCGA database.
Materials and Methods
Patient and Tissue Samples
In this study, a total of 94 ESCC samples were collected from TCGA (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga). RNA-seq expression profile data and survival data were downloaded from TCGA. What's more, RNA-seq expression profile data of 516 normal esophageal mucosa and muscle layers from GTEx database were downloaded from cancer genomics analysis platform from cancer genomics analysis platform from University of California Santa Cruz (UCSC Xena, http://xena.ucsc.edu/) (Deadline: February 14, 2023).
This study included 179 patients with ESCC from January 2010 to December 2017 in Affiliated Hospital 2 of Nantong University, of which 52 patients could obtain adjacent tissues of ESCC. Inclusion of samples should meet the following requirements: ESCC must be confirmed by pathology, the primary tumor was first diagnosed without anti-tumor treatment such as radiotherapy and chemotherapy before operation. And exclusion criteria included incomplete clinical data, loss to follow-up, and combination with other tumors. The final included cases were re-read by three qualified pathologists, and pTNM stage was carried out by UICC/AJCC stage system (8th edition, 2017). Overall survival (OS) is the time between the date of operation and the date of all-cause death or date of follow-up. Follow-up was performed by telephone until June 30, 2022.
Tissue Microarray and Immunohistochemistry
Tissue microarray was made from 2 mm tumor core and normal esophageal tissue from paraffin-embedded specimens fixed by formalin and then immunohistochemical staining was performed. After deparaffinization and hydration, antigen repair was promoted by heating in EDTA solution. The arrays were then probed with rabbit anti-human BRD4 (dilute 1: 400; ab128874, Abcom) and rabbit anti-human CD8 (dilute 1: 100; ab237709, Abcom). DAB coloration was followed by counterstained with hematoxylin.
The stained sections were interpreted and scored by three qualified pathologists. Negative is scored as 0. Light yellow, which means weakly positive, is recorded as 1 point. Brown yellow means moderately positive, which is recorded as 2 points. Dark brown, meaning strongly positive, is recorded as 3 points. Four 400 times visual fields were scored and then average value was taken (H-score = % unstained*0 + % weak staining*1 + % moderate staining*2 + %strong staining*3, score range 0 ~ 300) [11].
ESTIMATE Database
ESTIMATE (https://bioinformatics.mdanderson.org/estimate/) provides scores of tumor purity, presence level of stromal cells and infiltration levels of immune cells in tumor tissues according to expression data.
TIMER Database
TIMER 2.0 (http://timer.cistrome.org/) is a comprehensive resource for systematically analyzing immune infiltration of different cancer types which provides abundance of immune infiltration estimated by various immune deconvolution methods. CIBERSORT method was used to calculate the immune infiltration scores of ESCC in TCGA database [13, 14].
Statistical Analysis
SPSS 26.0, GraphPad Prism 8 and R software 4.0.5 were used for statistical analysis. R.utils, rjson and XML packages were used to read the raw data downloaded from TCGA database. Survival, dplyr and survminer packages were used to find the best cut-point value for grouping and perform Kaplan–Meier curve. Independent sample t test or rank sum test was adopted for comparison between measurement data groups, and χ2 test or Fisher exact test was used for comparison between counting data groups. The evaluation of prognostic value of BRD4 was performed by univariate and multivariate cox regression. Spearman and Phi coefficient were used for correlation analysis, while TIDE algorithm was used to predict treatment response to immune checkpoint blockade (ICB). Double tail P < 0.05 considered that the difference was statistically significant.
Results
Up-regulation of BRD4 Expression Level in ESCC
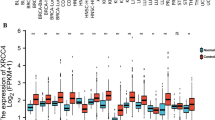
Firstly, this study compared the expression differences between ESCC and normal esophageal tissues of BET family members in the database. The expressions of BRD2, BRD3, BRD4 and BRDT in ESCC were higher than those in normal esophageal tissues (Fig. 1A), among which BRD2 and BRD4 accounted for the highest (Fig. 1B). After that, tissue microarray was used to verify the expression level of BRD4, which was expressed in the nucleus of cells, showing light yellow, brown yellow and dark brown according to the expression intensity. The results according to the statistics of the previous scoring system indicated that the expression level of BRD4 in ESCC was also up-regulated (Fig. 1C-D).
BRD4 is up-regulated in ESCC. a Expression level of BET family in ESCC and normal esophageal tissues in TCGA and GTEx databases. b Differential expression level of BET family in ESCC in TCGA database. c Expression level of BRD4 in ESCC tumor tissues and adjacent tissues in tissue microarray. d Typical immunohistochemical staining of ESCC tumor tissues and adjacent tissues in tissue microarray. ns P ≥ 0.05, ***P < 0.001
BRD4 High Expression Level Correlates with Poor Prognosis
To explore the correlations between BRD2/BRD4 and prognosis, this study has derived survival data from the TCGA cohort. To start with, Kaplan–Meier curve was performed to determine the best cut-off value related to OS, by which BRD4 low expression level group and BRD4 high expression level group were divided. According to the analysis, the OS of BRD4 high expression level group was shorter (Fig. 2A). Similarly, Kaplan–Meier curve of our cohort showed that the OS of BRD4 high expression level group was shorter (Fig. 2B). The above results suggested that BRD4 high expression level correlated to poor prognosis. However, there was no significant correlation between BRD2 expression level and survival (Fig. 2C). Therefore, the further study put a main focus on BRD4.
Univariate and multivariate cox regression were conducted on our cohort next. Univariate analysis showed that lymph node metastasis (P < 0.001), highly stage (P = 0. 003), intravascular tumor thrombus (P < 0.001) and high expression level of BRD4 (P = 0. 008) were risk factors for OS in ESCC (Table 1). Multivariate analysis included indicators whose P value was less than 0.2, and showed that lymph node metastasis (P = 0. 012) and intravascular tumor thrombus (P < 0.001) were the risk factors of OS in ESCC. However, BRD4 that the study was interested in was not statistically significant in multivariate analysis (P = 0. 081) (Table 1). To sum up, we believe that the high expression level of BRD4 is related to the poor prognosis of ESCC.
High Expression Level of BRD4 Is Associated with Adverse Clinicopathological Features
In order to explore which biological behaviors resulted in adverse prognostic outcomes are associated with BRD4, this study further analyzed the correlations between BRD4 and clinicopathological features. Likewise, BRD4 high expression level group and BED4 low expression level group were divided according to the best cut-off point determined by Kaplan–Meier survival curve. Our cohort analysis results showed that the high expression level of BRD4 was irrelevant to tumor occurrence sites, M stage, perineural invasion and intravascular tumor thrombus, but was related to T stage (P = 0. 001), N stage (P = 0. 034), clinical stage (P = 0. 005) and differentiation degree (P = 0. 020) (Table 2), indicating that the high expression level of BRD4 is related to adverse clinicopathological features and can affect the occurrence and development of ESCC.
BRD4 Expression Level Correlates with Systemic Inflammatory Response in ESCC
Next, the study collected the blood indexes of cohort cases one week before operation. Studies have shown that preoperative peripheral blood monocyte count, lymphocyte count, systemic inflammatory-immunologic index (SII, SII = platelet*neutrophil/lymphocyte), PLR (platelet/lymphocyte), MLR (monocyte/lymphocyte), NLR (neutrophil/lymphocyte) were related to tumor prognosis [15,16,17,18,19,20]. The grouping method was the same as above, and differences of the above six indexes between the two groups were compared. The results showed that the monocyte count (P = 0. 006), SII (P = 0. 015), PLR (P = 0. 042) and MLR (P = 0. 010) in the high expression level group of BRD4 were higher than those in the low expression level group (Fig. 3), indicating that BRD4 might regulate the systemic inflammatory response of patients with ESCC.
BRD4 expression level and systemic inflammatory response markers. a BRD4 expression level and total monocyte count. b BRD4 expression level and total lymphocytes count. c BRD4 expression level and SII. d BRD4 expression level and PLR. e BRD4 expression level and MLR. f BRD4 expression level and NLR. ns P ≥ 0.05, *P < 0.05, **P < 0.01
BRD4 Expression Level Correlates with Immune Infiltration in ESCC
In order to further explore the correlations between BRD4 and immune infiltration in ESCC, the study analyzed the TCGA ESCC cohort. Firstly, the data were imported into ESTIMATE website. And then, stromal, immune and ESTIMATE score were performed. The results showed that the immune score of BRD4 high expression level group was lower than that of BRD4 low expression level group (P = 0. 02) (Fig. 4A). Next, the immune infiltration scores of ESCC in TCGA ESCC cohort were calculated by CIBERSORT method on TIMER 2.0 website. The results indicated that the scores of CD8 + T cells and neutrophils were lower, and the scores of CD4 + memory resting T cells and myeloid dendritic resting cells were higher in the BRD4 high expression level group (Fig. 4B). We then examined the expression of CD8 in ESCC tissues in tissue microarray, which was mainly expressed in the cytoplasm. The Phi coefficient was -0.337, indicating a negative correlation between BRD4 and CD8, which was consistent with the results of TCGA database (Fig. 4C, Table 3).
The High Expression Level of BRD4 Correlated with the Low Response Rate of Immunotherapy
In the study, common genes related to immune escape and chemokines, interleukins and interferons in tumor microenvironment were collected [21]. The correlation analysis between above genes and BRD4 was conducted in TCGA ESCC cohort. The results indicated that BRD4 is positively correlated with related molecules promoting immune escape [22, 23], such as TGFB1 and STAT3. However, it was negatively correlated with molecules related to killing tumor cells [24], such as CD8B (Fig. 5A-B). Finally, we used TIDE algorithm to predict ICB response, and the results showed that both in TCGA esophageal cancer and ESCC cohorts, the BRD4 high expression level group had a higher score than the low expression level group (Fig. 5C-D).
BRD4 expression level and response to immunotherapy. a Correlation analysis between BRD4 expression level and immune escape related genes (Spearman). b Correlation analysis between BRD4 expression level and chemokines, interleukins and interferons in tumor microenvironment (Spearman). c Prediction of BRD4 expression level and the response to ICB in esophageal cancer by the TIDE algorithm. d Prediction of BRD4 expression level and the response to ICB in ESCC by the TIDE algorithm. **P < 0.01, ***P < 0.001
Discussion
According to the statistical report of GLOBOCAN in 2020, the number of new cases of esophageal cancer ranked seventh among 36 cancers in the world, and the number of fatal cases ranked sixth [25]. At present, the treatment of esophageal cancer mainly includes surgical resection, radiotherapy, chemotherapy, molecular targeted therapy and immunotherapy. In recent years, higher importance has been attached to immunotherapy. Besides, tumor vaccines and checkpoint inhibitors have also been used in the treatment of esophageal cancer. Compared with esophageal adenocarcinoma, ESCC is more suitable for immunotherapy due to its biological characteristics [26]. At present, pembrolizumab and Nivolumab have been approved by the US Food and Drug Administration (FAD) for the treatment of esophageal cancer [27]. Although more advanced treatment methods have been adopted, most patients with ESCC still suffered from poor prognosis. Therefore, it is of great urgency to find new biomarkers, carry out risk stratification of patients, conduct timely and accurately adjuvant treatment for high-risk patients.
BRD4 is an important member of the BET family. BRD4 domain plays a significant role in gene transcription, DNA replication and repair by binding with transcription regulators, and is related to the occurrence and development of various human diseases [28]. A large number of studies have shown that BRD4 is up-regulated in a variety of tumors, including non-small cell lung cancer, ovarian cancer, multiple myeloma, gastric cancer, colorectal cancer and prostate cancer [29,30,31,32,33,34]. This study observed that BRD4 was highly expressed in ESCC, which was consistent with previous studies. In addition, previous studies have shown that BRD4 can be used as a potential biomarker for evaluating the prognosis of tumors such as breast cancer, liver cancer, and bladder cancer [8,9,10]. This study also found that high expression level of BRD4 was associated with poor prognosis of ESCC. Besides, univariate and multivariate cox regression showed that high expression level of BRD4 was a risk factor for OS of ESCC in univariate analysis. However, BRD4 that we focused on lacks statistical significance in the multiple analysis. Since the factors involved were in great quantity that the result might be affected by insufficient sample size. Therefore, BRD4 might also be a potential biomarker for evaluating the prognosis of ESCC.
To explore which biological behaviors resulted in adverse prognostic outcomes are associated with BRD4, the study further analyzed the correlations between BRD4 and clinicopathological features. The results of our cohort showed that the high expression level of BRD4 was uncorrelated to tumor location, M stage, perineural invasion and intravascular tumor thrombus, but was related to T stage, N stage, clinical stage and differentiation degree. A large number of studies have shown that preoperative inflammatory indicators in peripheral blood, including monocyte count, lymphocyte count, SII, PLR, NLR and MLR, can effectively evaluate the prognosis of tumor patients [15,16,17,18,19,20]. We tried to analyze whether the expression of BRD4 is related to the above six indexes in this study. The results showed that monocyte counts were higher in the BRD4 high expression group. Similarly, the SII, PLR and MLR index of the BRD4 high expression level group were also higher. Previous studies have shown that BRD4 can regulate the expression of immune checkpoint genes. Studies by Zhao et al. indicated that BRD4 can regulate the expression of B7-H3 in pancreatic cancer [35]. In addition, Studies by Kazumi Ebine et al. showed that the interaction between BRD4 and IRF1 can regulate the expression of PD-L1 in pancreatic cancer [36]. Therefore, we further analyzed the relationship between BRD4 and immune infiltration in ESCC.
In addition to tumor cells, solid tumors also contain special cellular and non-cellular components, which constitute the tumor microenvironment. The cellular components of tumor microenvironment mainly include neuroendocrine cells, adipocytes, endothelial cells, mesenchymal cells, immune inflammatory cells and fibroblasts [37]. The microenvironment also changes during tumor development. Initially, most of the infiltrating immune cells play an anti-tumor effect, but a small number of immune cells will promote the development of tumors, but the specific subtypes need to be further studied [38]. What is clear is that T cells play a suppressive role. Activated T cells are associated with a good prognosis in a variety of tumors [39]. After that, the infiltrating immune cells will gradually increase, but the immune cells with killing effect will gradually disappear, which may be the exhaustion of T cells, leading to immune escape of tumor cells [40].
We used the ESTIMATE website to perform stromal, immune, and ESTIMATE score for the TCGA ESCC cohort. The results showed that the immune score of the BRD4 high expression level group was lower than that of the low expression level group. Next, the immune infiltration scores in TCGA ESCC cohort were calculated using CIBERSORT on the TIMER 2.0 website. The results showed that BRD4 expression level was negatively correlated with the infiltration of CD8 + T cells, which is the killing immune cells. Similarly, the expression of CD8 in ESCC tissues was detected in tissue microarray, and the correlation analysis was performed with the expression level of BRD4, and there was a negative correlation between them. Subsequently, the correlation analysis between BRD4 and common immune escape genes and chemokines, interleukins and interferons in the tumor microenvironment was carried out. The results showed that BRD4 was positively correlated with molecules related to promoting immune escape [22, 23], such as TGFB1 and STAT3. However, it is negatively correlated with molecules related to killing tumor cells [24], such as CD8B. Therefore, patients with high BRD4 expression level may be more susceptible to immune escape.
Finally, the TIDE algorithm was used to predict the response to ICB, and the results showed that both in the TCGA esophageal cancer and ESCC cohorts, higher TIDE scores in the BRD4 high expression group than in the low expression group. Therefore, esophageal cancer patients with high BRD4 expression level may not be suitable for immunotherapy. When BRD4 is highly expressed, the combination of BRD4 inhibitors and immunotherapy may be a novel therapeutic strategy to improve the efficacy of patients with esophageal cancer.
In conclusion, BRD4 is up-regulated in ESCC. The high expression level of BRD4 in ESCC is associated with poor prognosis, adverse clinicopathological features, immune infiltration, and low response rate to immunotherapy. Therefore, BRD4 may be a potential biomarker for the prognosis and immunotherapy application of ESCC.
Data Availability
All data generated or analyzed during this study are included in this published article.
References
Abnet CC, Arnold M, Wei WQ. Epidemiology of Esophageal Squamous Cell Carcinoma. Gastroenterology. 2018;154:360–373. https://doi.org/10.1053/j.gastro.2017.08.023.
Yang J, Liu X, Cao S, Dong X, Rao S, Cai K. Understanding Esophageal Cancer: The Challenges and Opportunities for the Next Decade. Frontiers in oncology. 2020;10:1727. https://doi.org/10.3389/fonc.2020.01727.
Yang Y, Ge H. Effective combinations of radiotherapy and immunotherapy in the treatment of esophageal squamous cell carcinoma. Future oncology (London, England). 2020;16:2537–2549. https://doi.org/10.2217/fon-2020-0222.
Dai E, Zhu Z, Wahed S, Qu Z, Storkus WJ, Guo ZS. Epigenetic modulation of antitumor immunity for improved cancer immunotherapy. Molecular cancer. 2021;20:171. https://doi.org/10.1186/s12943-021-01464-x.
Ali HA, Li Y, Bilal AHM, Qin T, Yuan Z, Zhao W. A Comprehensive Review of BET Protein Biochemistry, Physiology, and Pathological Roles. Frontiers in pharmacology. 2022;13:818891. https://doi.org/10.3389/fphar.2022.818891.
Qin ZY, Wang T, Su S, Shen LT, Zhu GX, Liu Q et al. BRD4 Promotes Gastric Cancer Progression and Metastasis through Acetylation-Dependent Stabilization of Snail. Cancer research. 2019;79:4869–4881. https://doi.org/10.1158/0008-5472.Can-19-0442.
Wang N, Wu R, Tang D, Kang R. The BET family in immunity and disease. Signal transduction and targeted therapy. 2021;6:23. https://doi.org/10.1038/s41392-020-00384-4.
Zhang Y, Xu B, Shi J, Li J, Lu X, Xu L et al. BRD4 modulates vulnerability of triple-negative breast cancer to targeting of integrin-dependent signaling pathways. Cellular oncology (Dordrecht). 2020;43:1049–1066. https://doi.org/10.1007/s13402-020-00537-1.
Zhang P, Dong Z, Cai J, Zhang C, Shen Z, Ke A et al. BRD4 promotes tumor growth and epithelial-mesenchymal transition in hepatocellular carcinoma. International journal of immunopathology and pharmacology. 2015;28:36–44. https://doi.org/10.1177/0394632015572070.
Yan Y, Yang FQ, Zhang HM, Li J, Li W, Wang GC et al. Bromodomain 4 protein is a predictor of survival for urothelial carcinoma of bladder. International journal of clinical and experimental pathology. 2014;7:4231–4238.
Niu H, Song F, Wei H, Li Y, Huang H, Wu C. Inhibition of BRD4 Suppresses the Growth of Esophageal Squamous Cell Carcinoma. Cancer investigation. 2021;39:826–841. https://doi.org/10.1080/07357907.2021.1975736.
Wu Q, Liu F, Ge M, Laster KV, Wei L, Du R et al. BRD4 drives esophageal squamous cell carcinoma growth by promoting RCC2 expression. Oncogene. 2022;41:347–360. https://doi.org/10.1038/s41388-021-02099-4.
Floberg JM, Zhang J, Muhammad N, DeWees TA, Inkman M, Chen K et al. Standardized Uptake Value for (18)F-Fluorodeoxyglucose Is a Marker of Inflammatory State and Immune Infiltrate in Cervical Cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2021;27:4245–4255. https://doi.org/10.1158/1078-0432.Ccr-20-4450.
Xue G, Hua L, Zhou N, Li J. Characteristics of immune cell infiltration and associated diagnostic biomarkers in ulcerative colitis: results from bioinformatics analysis. Bioengineered. 2021;12:252–265. https://doi.org/10.1080/21655979.2020.1863016.
Feng F, Zheng G, Wang Q, Liu S, Liu Z, Xu G et al. Low lymphocyte count and high monocyte count predicts poor prognosis of gastric cancer. BMC gastroenterology. 2018;18:148. https://doi.org/10.1186/s12876-018-0877-9.
Zhang X, Hu D, Lin X, Zhang H, Xia Y, Lin J et al. Prognostic Value of an Inflammation-Related Index in 6,865 Chinese Patients With Postoperative Digestive Tract Cancers: The FIESTA Study. Frontiers in oncology. 2019;9:427. https://doi.org/10.3389/fonc.2019.00427.
Zhang Y, Lin S, Yang X, Wang R, Luo L. Prognostic value of pretreatment systemic immune-inflammation index in patients with gastrointestinal cancers. Journal of cellular physiology. 2019;234:5555–5563. https://doi.org/10.1002/jcp.27373.
Feng JF, Chen S, Yang X. Systemic immune-inflammation index (SII) is a useful prognostic indicator for patients with squamous cell carcinoma of the esophagus. Medicine. 2017;96:e5886. https://doi.org/10.1097/md.0000000000005886.
Chen H, Wu X, Wen Z, Zhu Y, Liao L, Yang J. The Clinicopathological and Prognostic Value of NLR, PLR and MLR in Non-Muscular Invasive Bladder Cancer. Archivos espanoles de urologia. 2022;75:467–71. https://doi.org/10.56434/j.arch.esp.urol.20227505.68.
Palacios-Acedo AL, Mège D, Crescence L, Dignat-George F, Dubois C, Panicot-Dubois L. Platelets, Thrombo-Inflammation, and Cancer: Collaborating With the Enemy. Frontiers in immunology. 2019;10:1805. https://doi.org/10.3389/fimmu.2019.01805.
Hu Z, Qu S. EVA1C Is a Potential Prognostic Biomarker and Correlated With Immune Infiltration Levels in WHO Grade II/III Glioma. Frontiers in immunology. 2021;12:683572. https://doi.org/10.3389/fimmu.2021.683572.
Zhang L, Kuca K, You L, Zhao Y, Musilek K, Nepovimova E, et al. Signal transducer and activator of transcription 3 signaling in tumor immune evasion. Pharmacology & therapeutics. 2022;230:107969. https://doi.org/10.1016/j.pharmthera.2021.107969.
Wang Z, Lu Z, Lin S, Xia J, Zhong Z, Xie Z et al. Leucine-tRNA-synthase-2-expressing B cells contribute to colorectal cancer immunoevasion. Immunity. 2022;55:1067–81.e8. https://doi.org/10.1016/j.immuni.2022.04.017.
Freed-Pastor WA, Lambert LJ, Ely ZA, Pattada NB, Bhutkar A, Eng G et al. The CD155/TIGIT axis promotes and maintains immune evasion in neoantigen-expressing pancreatic cancer. Cancer cell. 2021;39:1342–60.e14. https://doi.org/10.1016/j.ccell.2021.07.007.
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: a cancer journal for clinicians. 2021;71:209–49. https://doi.org/10.3322/caac.21660.
Mimura K, Yamada L, Ujiie D, Hayase S, Tada T, Hanayama H et al. Immunotherapy for esophageal squamous cell carcinoma: a review. Fukushima journal of medical science. 2018;64:46–53. https://doi.org/10.5387/fms.2018-09.
Kelly RJ, Ajani JA, Kuzdzal J, Zander T, Van Cutsem E, Piessen G et al. Adjuvant Nivolumab in Resected Esophageal or Gastroesophageal Junction Cancer. The New England journal of medicine. 2021;384:1191–1203. https://doi.org/10.1056/NEJMoa2032125.
Shi Y, Liu J, Zhao Y, Cao J, Li Y, Guo F. Bromodomain-Containing Protein 4: A Druggable Target. Current drug targets. 2019;20:1517–1536. https://doi.org/10.2174/1574885514666190618113519.
Fan H, Yuan J, Li Y, Jia Y, Li J, Wang X et al. MKL1-induced lncRNA SNHG18 drives the growth and metastasis of non-small cell lung cancer via the miR-211-5p/BRD4 axis. Cell death & disease. 2021;12:128. https://doi.org/10.1038/s41419-021-03399-z.
Zeng XY, Yuan J, Wang C, Zeng D, Yong JH, Jiang XY et al. circCELSR1 facilitates ovarian cancer proliferation and metastasis by sponging miR-598 to activate BRD4 signals. Molecular medicine (Cambridge, Mass). 2020;26:70. https://doi.org/10.1186/s10020-020-00194-y.
Zheng JF, Guo NH, Zi FM, Cheng J. Long Noncoding RNA H19 Promotes Tumorigenesis of Multiple Myeloma by Activating BRD4 Signaling by Targeting MicroRNA 152–3p. Molecular and cellular biology. 2020;40. https://doi.org/10.1128/mcb.00382-19.
Ba M, Long H, Yan Z, Wang S, Wu Y, Tu Y et al. BRD4 promotes gastric cancer progression through the transcriptional and epigenetic regulation of c-MYC. Journal of cellular biochemistry. 2018;119:973–982. https://doi.org/10.1002/jcb.26264.
Otto C, Schmidt S, Kastner C, Denk S, Kettler J, Müller N et al. Targeting bromodomain-containing protein 4 (BRD4) inhibits MYC expression in colorectal cancer cells. Neoplasia (New York, NY). 2019;21:1110–1120. https://doi.org/10.1016/j.neo.2019.10.003.
Tan Y, Wang L, Du Y, Liu X, Chen Z, Weng X et al. Inhibition of BRD4 suppresses tumor growth in prostate cancer via the enhancement of FOXO1 expression. International journal of oncology. 2018;53:2503–2517. https://doi.org/10.3892/ijo.2018.4577.
Zhao J, Meng Z, Xie C, Yang C, Liu Z, Wu S et al. B7–H3 is regulated by BRD4 and promotes TLR4 expression in pancreatic ductal adenocarcinoma. The international journal of biochemistry & cell biology. 2019;108:84–91. https://doi.org/10.1016/j.biocel.2019.01.011.
Ebine K, Kumar K, Pham TN, Shields MA, Collier KA, Shang M et al. Interplay between interferon regulatory factor 1 and BRD4 in the regulation of PD-L1 in pancreatic stellate cells. Scientific reports. 2018;8:13225. https://doi.org/10.1038/s41598-018-31658-1.
Fiori ME, Di Franco S, Villanova L, Bianca P, Stassi G, De Maria R. Cancer-associated fibroblasts as abettors of tumor progression at the crossroads of EMT and therapy resistance. Molecular cancer. 2019;18:70. https://doi.org/10.1186/s12943-019-0994-2.
Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad M et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nature medicine. 2018;24:541–550. https://doi.org/10.1038/s41591-018-0014-x.
Gajewski TF, Corrales L, Williams J, Horton B, Sivan A, Spranger S. Cancer Immunotherapy Targets Based on Understanding the T Cell-Inflamed Versus Non-T Cell-Inflamed Tumor Microenvironment. Advances in experimental medicine and biology. 2017;1036:19–31. https://doi.org/10.1007/978-3-319-67577-0_2.
Wang JC, Xu Y, Huang ZM, Lu XJ. T cell exhaustion in cancer: Mechanisms and clinical implications. Journal of cellular biochemistry. 2018;119:4279–4286. https://doi.org/10.1002/jcb.26645.
Funding
This study was supported by Postgraduate Research & Practice Innovation Program of Jiangsu Province (SJCX21_1469) and Nantong Science and Technology Project (JC2021001).
Author information
Authors and Affiliations
Contributions
HL designed the study. LL and LG performed the research and wrote the paper. HZ, CS and XZ staged the pTNM stage of patients and scored the results of immunohistochemical staining. DZ analyzed data.
Corresponding author
Ethics declarations
Conflict of interest
The Authors declare that they have no conflict of interest.
Ethical approval
The Investigation conformed to the principles outlined in the Declaration of Helsinki. This study was conducted under the approval of the appropriate Ethics Committees in Affiliated Hospital 2 of Nantong University (IRB No.: 2021KT057).
Informed consent
Written informed consent was obtained for each participant.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc/4.0/.
About this article
Cite this article
Li, L., Gao, L., Zhou, H. et al. High Expression Level of BRD4 Is Associated with a Poor Prognosis and Immune Infiltration in Esophageal Squamous Cell Carcinoma. Dig Dis Sci 68, 2997–3008 (2023). https://doi.org/10.1007/s10620-023-07907-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10620-023-07907-3