Abstract
Background
Computed tomography is the most commonly used imaging modality for preoperative assessment of lymph node status, but the reported accuracy is unsatisfactory.
Aims
To evaluate and verify the predictive performance of computed tomography deep learning on the presurgical evaluation of lymph node metastasis in patients with gastric cancer.
Methods
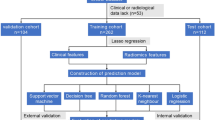
347 patients were retrospectively selected (training cohort: 242, test cohort: 105). The enhanced computed tomography arterial phase images of gastric cancer were used for lesion segmentation, radiomics and deep learning feature extraction. Three methods were used for feature selection. Support vector machine (SVM) or random forest (RF) was used to build models. The classification performance of the models was evaluated using the area under the receiver operating characteristic curve (AUC). We also established a nomogram that included clinical predictors.
Results
The model based on ResNet50-RF showed favorable classification performance and was verified in the test cohort (AUC = 0.9803). The nomogram based on deep learning feature scores and the lymph node status reported by computed tomography showed excellent discrimination. AUC of 0.9978 was achieved in the training cohort and verified in the test cohort (AUC = 0.9914). Decision analysis curve showed the value of nomogram in clinical application.
Conclusion
The computed tomography-based deep learning nomogram can accurately and effectively evaluate lymph node metastasis in patients with gastric cancer before surgery.
Similar content being viewed by others
Explore related subjects
Find the latest articles, discoveries, and news in related topics.Avoid common mistakes on your manuscript.
Introduction
Gastric cancer is an aggressive malignant tumor with a poor prognosis and the fifth most common malignant tumor in the world [1]. Because the early symptoms of gastric cancer are not typical, many patients have developed advanced gastric cancer at the time of diagnosis [2]. The main way of gastric cancer metastasis is lymphatic metastasis, which affects the prognosis of gastric cancer [3, 4]. The presence or absence of lymph node (LN) metastasis is one of the criteria for neoadjuvant chemotherapy, which is of great importance for patient's prognosis with gastric cancer and the choice of chemotherapy regimens [4,5,6]. Therefore, it is of great importance for patient treatment and improving prognosis to diagnose LN metastasis. The most common method for preoperative assessment of LN metastasis is computed tomography (CT) [7]. The accepted criteria for clinical diagnosis of LN metastasis are changes in size and shape [8]. However, its accuracy is not satisfactory, and some patients are at risk of inaccurate LN staging [9,10,11]. Thus, a non-invasive method is needed to assist imaging diagnosis.
Recently, artificial intelligence (AI) is an emerging technology that provides new methods for assisted diagnosis of medical imaging. Studies have confirmed that the accuracy of diagnosis using AI models is similar to or even better than that of clinical experts under certain conditions [12,13,14]. Transfer learning can overcome the limitations of small data sets [15]. It uses pre-trained convolutional neural network (CNN) for deep learning (DL) feature extraction and builds models with the help of machine learning (ML) methods [16]. There have been many studies on radiomics in the diagnosis and prognosis of tumors [17, 18]. However, there are few studies using deep learning to predict LN metastasis in gastric cancer [19, 20].
Therefore, this study aims to develop a non-invasive method based on deep learning to predict gastric cancer LN metastasis before surgery and combine it with clinical predictors to construct a nomogram.
Materials and Methods
We obtained informed consent from patients or their relatives and were approved by the Ethics Committee of the Second Affiliated Hospital of Nanjing Medical University (NO. [2022]-KY-009-01). All patient private information was deleted.
Quality System
The study followed the RQS quality system. The RQS scoring criteria, the scores for this study and the rationale for the scores were detailed in the supplementary material.
Patients
We reviewed patients from January 2017 to September 2021. Inclusion criteria included abdominal enhanced CT examination within one week before surgery and no neoadjuvant chemotherapy, postoperative pathological confirmation of gastric cancer and clear staging. Exclusion criteria include poor CT image quality and small gastric cancer lesions that are difficult to identify. We collected the information of patients who met the inclusion criteria, and then excluded patients who met the exclusion criteria. In the end, we selected 347 patients (mean age, 64.34 ± 11.075 years; the range of the ages, 29–90 years) for review. Clinical, pathological and laboratory data all came from medical records. The CT image acquisition was detailed in supplementary materials. Three radiologists (with more than 7 years of experience in abdominal imaging) reviewed the patient's enhanced CT arterial phase images and reassessed the patient's LN status. The evaluation processes of the three doctors were independent of each other, and they had no knowledge of the patient's pathological information. If the opinions of the three doctors are not uniform, we will take the majority opinion as the patient’s final LN status.
Regions of Interests (ROIs)
Studies have shown that the predictive performance of radiomic features extracted from CT images of the arterial phase is better than that of the portal phase [21, 22]. Thus, we resampled all CT arterial phase images. Two imaging physicians (with more than 7 years of work experience) used ITK-SNAP software to semi-automatically segment the gastric cancer ROI in the arterial phase of CT images. When performing gastric cancer ROI segmentation, the doctors only knew the location of the tumor confirmed by pathology and did not know the rest of the information. Using the CT attenuation threshold, a semi-automatic method was used to delineate free regions of interest along the tumor margins. Top and bottom slices were excluded to reduce bias due to local volume effects. The radiologist will carefully outline the contours to avoid contact with adjacent fluid or air, and areas of necrosis will be included.
Radiomics Features
We extracted radiomics features from the ROI with the help of the PyRadiomics package (version 3.0.1) [23]. Most features were in line with the image biomarker standardization initiative [24, 25]. We use the intraclass correlation coefficient (ICC) with the help of the pingouin package (version 0.3.11) to verify feature stability and select features with a value greater than 0.8 [26, 27]. Feature extraction for radiomics and calculation of ICC were detailed in supplementary materials.
Deep Learning Features
VGG16, VGG19, Xception, InceptionV3, and InceptionResNetV2 were used to extract deep learning features. We also used a total of five different layers of ResNet50 for deep learning feature extraction. All neural networks were pre-trained on the ImageNet database. The extracted deep learning features were modeled by ML methods. Deep learning was less dependent on the understanding of data features [28]. The training process of the CNN and the extraction of deep learning features were detailed in the supplementary materials.
Feature Selection and Model Construction
To deal with the imbalance of the LN status, we use the Borderline-SMOTE method to oversample the training cohort. Due to the large number of features, we normalized the feature value and followed the steps below to perform feature selecting. First, we used T-test for preliminary screening and selected features with p < 0.05. Then, we used the method of univariate analysis to select the top 20% of the best features. Finally, we used the least absolute shrinkage and selection operator (Lasso) method to select features and recorded the feature scores of all patients. We also integrated the selected deep learning features and radiomics features, and standardized and selected these features again. The specific methods were detailed in the supplementary materials. Mann–Whitney U test was used to verify the association between features and LN status. We built models for the selected six sets of deep learning features, radiomics features and integrated features. Since the classification performance of models built with different classifiers may have obvious differences, we used SVM or RF to build classification models, and used the AUC to evaluate model performance.
Nomogram Construction
All the clinical, pathological and laboratory data, including age, gender, tumor location, tumor morphology, album, neutrophil, lymphocyte, CEA level, CA742 level, CT-reported LN status, and deep learning feature scores were evaluated with univariate analysis and logistic regression analysis. We used likelihood ratio test to select factors related to LN metastasis. On the basis of the results of the above analysis, a nomogram was constructed.
Statistical Analysis
The Kolmogorov–Smirnov test was used to test the normality of continuous variables. Differences between normally distributed variables were compared with T test. When the variables were non-normally distributed, the Mann–Whitney U test was used to analyze the differences between these variables. Categorical variables were expressed by frequency. Chi-square test was used to test their differences. The violin graph was used to represent the distribution of the deep learning feature scores of each cohort. The performance of the classification models was evaluated by the AUC. Calibration curve and Hosmer–Lemeshow test were used to evaluate the nomogram. Receiver operator characteristics (ROC) curves were used to evaluate the predictive performance of the nomogram and the diffidence of them were compared by Delong test. Decision analysis curve (DCA) was drawn to showed the value of the classification model and nomogram in clinical application [29].
Results
Clinical Characteristics
Among the 347 patients who met the inclusion criteria of this study, 242 patients were randomly divided into the training cohort, and the remaining 105 patients were divided into the test cohort randomly. Table 1 summarized the clinical characteristics of each cohort. The clinical characteristics of the two cohorts did not differ significantly.
Model Construction and Evaluation
After a series of feature selecting, we selected 33 features from VGG16, 39 features from VGG19, 69 features from Xception, 85 features from InceptionV3, 77 features from InceptionResNetV2 and 72 features from ResNet50. We also selected 26 radiomics features (Fig. 1).
Among the models built using deep learning features, the ResNet50-RF model has the optimal classification performance. Its AUC, accuracy, sensitivity and specificity were 0.9803, 0.9810, 0.9839, 0.9767, respectively (Table S1). We also analyzed and compared the features extracted from different layers of ResNet50, and the results confirmed that the last layer before the neural network was fully connected to extract features was the most effective (Table S2).
We used the selected radiomics features to build model. The classification models on the basis of RF or SVM showed similar performance (DeLong test, p = 0.30728). Its AUC, accuracy, sensitivity and specificity were 0.9606, 0.9619, 0.9677, 0.9535, respectively (Table S1).
We had also integrated the selected deep learning features extracted by ResNet50 and radiomics features to explore whether this method was feasible to improve classification performance. Analysis showed that integrating features can not improve classification performance (Table S1). The AUC of the Int-RF model was 0.9687.
The analysis results showed that the ResNet50-RF model had the best classification performance. The distribution of deep learning feature scores is shown in Fig. 2. In both cohorts, the deep learning feature scores of patients with positive LN were significantly higher than those with negative LN. Mann–Whitney U test (p < 0.001) confirmed that there was a correlation between deep learning features and LN status.
Nomogram Construction and Evaluation
The results of univariate analysis and logistic regression analysis showed that the deep learning feature scores and the LN status reported by CT were independent factors (Table 2). The nomogram (Fig. 3) on the basis of the two factors showed excellent predictive performance (Hosmer–Lemeshow test, p = 0.291). The AUC (Fig. 4) was 0.9978 in the train cohort, which was verified in the test cohort (AUC = 0.9914). The calibration curves indicated that the deep learning nomogram had excellent predictive performance (Fig. 5).
Deep learning nomogram. The deep learning nomogram was constructed in the training cohort, including deep learning feature scores and the LN status reported by CT. Image: The LN status reported by CT. Score: The deep learning feature scores. 0: Lymph node metastasis negative; 1: Lymph node metastasis positive
Calibration curves of the deep learning nomogram in each cohort. A Training cohort. B Test cohort. Dashed lines indicate perfect predictions. The prediction performances of the deep learning nomogram are represented by solid lines. The solid line and the dashed line are very close, which indicates that the deep learning nomogram has excellent predictive performance
The DCA of the classification model and nomogram are shown in Fig. 6. The analysis of the results showed that within a certain threshold range, the use of nomogram and classification model to predict whether a patient’s LN metastasis was more beneficial than treat-all patients or treat-none scheme.
Discussion
This study constructed a non-invasive method, CT-based deep learning nomogram for the preoperative evaluation of gastric cancer LN status, verified by the test cohort. The nomogram included two factors: deep learning feature scores and LN status reported by CT. The nomogram provided more information for the preoperative evaluation of LN.
In clinical work, medical imaging is a vital method to assess the status of LN [30]. CT is the most commonly used method [7]. However, the accuracy of CT in assessing LN metastasis is very limited [9,10,11, 31]. Radiomics is a new technology, which shows great potential in oncology [32,33,34]. Deep learning can automatically learn relevant features and reduce the amount of human input [35]. But classification using deep learning is difficult to combine with radiomics. In the field of gastric cancer imaging, research in recent years has only focused on radiomics or deep learning, and there is no article combining the two for research [17, 19, 36].
Therefore, there were two aspects of this problem have to be addressed. Firstly, we used CNN for deep learning feature extraction. However, there was no clear report in the recent study for which layer of CNN output depth features were used. Thus, we used the five different layers to output deep learning features of ResNet50 to build classification models for analysis and comparison. The results showed that the last layer before the neural network was fully connected to extract features was the most effective, which will serve as a reference for other researchers. Secondly, due to the large number of features, we used three methods for feature selecting. The analysis confirmed that the performance of the classification model we constructed using T-test, univariate analysis and Lasso method for feature selecting outperformed the models constructed by Chen et al. [17] (AUC, 0.9803 vs 0.878) and Wang et al. [36] (AUC, 0.9803 vs 0.837) using single feature selecting method.
Then, we combined the deep learning features and the radiomics features to build a classification model. But surprisingly, after combining the two, the performance of the classification model did not improve, which is similar to the study by Yun et al. [37] Chalkidou et al. [38] believed that radiomic features were characterized by human bias. At the same time, there had always been a problem of reproducibility in radiomics [39]. We were inclined to this view. Human-defined radiomics had certain limitations, and the differences between tissue types may not be fully included in the radiomic features.
Besides, we combined feature scores and clinical factors to build nomogram. Both clinicians and patients can use this easy-to-use nomogram, which is in line with the trend of personalized medicine [40]. Wang et al. [36] constructed a nomogram based on radiomics scores for preoperative assessment of the status of gastric cancer LN. In comparison, the nomogram of this study achieved a higher AUC (0.9914 VS 0.881). The better performance of this research may be due to the inclusion of deep learning feature scores. The advances of deep learning can effectively deal with the dilemmas of radiomics mentioned above. Deep learning can automatically learn features without human pre-definition, thereby reducing human bias and improving versatility and accuracy [35]. The nomogram had important guiding significance in helping clinicians to screen patients receiving neoadjuvant chemotherapy. Evidence-based evidence for the survival benefit of neoadjuvant chemotherapy still did not exist. Some patients with gastric cancer that could be cured by surgery may have serious adverse reactions after neoadjuvant chemotherapy, which was unnecessary. Accurate prediction of LN metastasis was the key to screening patients receiving neoadjuvant chemotherapy [4,5,6]. Therefore, to evaluate the clinical applicability of classification model and nomogram, this study adopted a DCA. Analysis showed that within a certain threshold, compared with treat-all-patients or treat-none scheme, using classification models or nomograms to predict LN metastasis can provide net benefits.
This study had some notable limitations. First of all, the deep learning features were obtained using CNN for transfer learning. Although the test cohort verification proved that the classification model had good predictive performance, there were differences between the source database and the target database. One solution is to build a large database that includes a large number of well-annotated medical imaging data. With the help of this database for CNN training, better performance will be obtained. Secondly, this study did not consider genomic characteristics. In recent years, radiogenomics has gained more and more attention in the field of oncology. Perhaps this is a worthwhile attempt, but it remains to be determined whether it is better to build a model that uses imaging features to directly predict the results than deep learning or radiomic analysis [41]. In addition, this study only focused on the presence or absence of LN metastasis. The role of deep learning and radiomics on the detailed N staging of patients and the specific grouping of LN deserved further study. Finally, this study had a small amount of data and was a retrospective study. Further studies need more data and external verification.
In conclusion, a nomogram was constructed and verified based on deep learning feature scores and clinical risk factor in the study and could use to assist individualized prediction of preoperative LN metastasis in patients with gastric cancer.
Data availability
The data supporting this study are available on request from the corresponding author.
References
Sung H, Ferlay J, Siegel R et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71:209–249.
Rice TW, Gress DM, Patil DT, Hofstetter WL, Kelsen DP, Blackstone EH. Cancer of the esophagus and esophagogastric junction-Major changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA Cancer J Clin 2017;67(4):304–317.
Zhou YX, Yang LP, Wang ZX et al. Lymph node staging systems in patients with gastric cancer treated with D2 resection plus adjuvant chemotherapy. J Cancer 2018;9:660–666.
Fukagawa T, Katai H, Mizusawa J et al. A prospective multi-institutional validity study to evaluate the accuracy of clinical diagnosis of pathological stage III gastric cancer (JCOG1302A). Gastric Cancer 2018;21:68–73.
Smyth EC, Verheij M, Allum W, Cunningham D, Cervantes A, Arnold D. Gastric cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2016;27:v38–v49.
Ajani JA, D’Amico TA, Almhanna K et al. Gastric Cancer, Version 3.2016, NCCN Clinical Practice Guidelines in oncology. J Natl Compr Cancer Netw 2016;14:1286–1312.
Li J, Fang M. Wang R et al. Diagnostic accuracy of dual-energy CT-based nomograms to predict lymph node metastasis in gastric cancer. Eur Radiol 2018;28:5241–5249.
In H, Solsky I, Palis B, Langdon-Embry M, Ajani J, Sano T. Validation of the 8th Edition of the AJCC TNM staging system for gastric cancer using the national cancer database. Ann Surg Oncol 2017;24(12):3683–3691.
Kim AY, Kim HJ, Ha HK. Gastric cancer by multidetector row CT: preoperative staging. Abdominal Imaging 2005;30:465–472.
Kinner S, Maderwald S, Albert J et al. Discrimination of benign and malignant lymph nodes at 7.0T compared to 1.5T magnetic resonance imaging using ultrasmall particles of iron oxide: a feasibility preclinical study. Acad Radiol 2013;20:1604–1609.
Kim HJ, Kim AY, Oh ST et al. Gastric cancer staging at multi-detector row CT gastrography: comparison of transverse and volumetric CT scanning. Radiology 2005;236:879–885.
Arribas J, Antonelli G, Frazzoni L et al. Standalone performance of artificial intelligence for upper GI neoplasia: a meta-analysis. Gut 2020.
Jiang K, Jiang X, Pan J et al. Current evidence and future perspective of accuracy of artificial intelligence application for early gastric cancer diagnosis with endoscopy: a systematic and meta-analysis. Front Med (Lausanne) 2021;8:629080.
Kermany DS, Goldbaum M, Cai W et al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell 2018;172:1122-1131.e9.
Shin HC, Roth HR, Gao M et al. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans Med Imaging 2016;35:1285–1298.
Raghu S, Sriraam N, Temel Y, Rao SV, Kubben PL. EEG based multi-class seizure type classification using convolutional neural network and transfer learning. Neural Netw 2020;124:202–212.
Chen W, Wang S, Dong D et al. Evaluation of lymph node metastasis in advanced gastric cancer using magnetic resonance imaging-based radiomics. Front Oncol 2019;9:1265.
Li W, Zhang L, Tian C et al. Prognostic value of computed tomography radiomics features in patients with gastric cancer following curative resection. Eur Radiol 2019;29:3079–3089.
Gao Y, Zhang ZD, Li S et al. Deep neural network-assisted computed tomography diagnosis of metastatic lymph nodes from gastric cancer. Chin Med J (Engl) 2019;132:2804–2811.
Bollschweiler EH, Monig SP, Hensler K, Baldus SE, Maruyama K, Holscher AH. Artificial neural network for prediction of lymph node metastases in gastric cancer: a phase II diagnostic study. Ann Surg Oncol 2004;11:506–511.
Ba-Ssalamah A, Muin D, Schernthaner R et al. Texture-based classification of different gastric tumors at contrast-enhanced CT. Eur J Radiol 2013;82:e537–e543.
Liu S, Liu S, Ji C et al. Application of CT texture analysis in predicting histopathological characteristics of gastric cancers. Eur Radiol 2017;27:4951–4959.
van Griethuysen JJM, Fedorov A, Parmar C et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res 2017;77:e104–e107.
Zwanenburg A, Leger S, Agolli L et al. Assessing robustness of radiomic features by image perturbation. Sci Rep 2019;9:614.
Balagurunathan Y, Kumar V, Gu Y et al. Test-retest reproducibility analysis of lung CT image features. J Digit Imaging 2014;27:805–823.
Aerts HJ, Velazquez ER, Leijenaar RT et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014;5:4006.
Yuan M, Zhang YD, Pu XH et al. Comparison of a radiomic biomarker with volumetric analysis for decoding tumour phenotypes of lung adenocarcinoma with different disease-specific survival. Eur Radiol 2017;27:4857–4865.
Huang S, Yang J, Fong S, Zhao Q. Artificial intelligence in cancer diagnosis and prognosis: opportunities and challenges. Cancer Lett 2020;471:61–71.
Vickers AJ, Cronin AM, Elkin EB, Gonen M. Extensions to decision curve analysis, a novel method for evaluating diagnostic tests, prediction models and molecular markers. BMC Med Inform Decis Mak 2008;8:53.
Giganti F, Tang L, Baba H. Gastric cancer and imaging biomarkers: Part 1—a critical review of DW-MRI and CE-MDCT findings. Eur Radiol 2019;29:1743–1753.
Cardoso R, Coburn N, Seevaratnam R et al. A systematic review and meta-analysis of the utility of EUS for preoperative staging for gastric cancer. Gastric Cancer 2012;15:S19-26.
Kumar V, Gu Y, Basu S et al. Radiomics: the process and the challenges. Magn Reson Imaging 2012;30:1234–1248.
Lambin P, Rios-Velazquez E, Leijenaar R et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer (Oxford, England: 1990) 2012;48(4):441–446.
Lambin P, Leijenaar RTH, Deist TM et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 2017;14:749–762.
LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015;521:436–444.
Wang Y, Liu W, Yu Y et al. CT radiomics nomogram for the preoperative prediction of lymph node metastasis in gastric cancer. Eur Radiol 2020;30:976–986.
Yun J, Park JE, Lee H, Ham S, Kim N, Kim HS. Radiomic features and multilayer perceptron network classifier: a robust MRI classification strategy for distinguishing glioblastoma from primary central nervous system lymphoma. Sci Rep 2019;9:5746.
Chalkidou A, O’Doherty MJ, Marsden PK. False discovery rates in PET and CT studies with texture features: a systematic review. PLoS ONE 2015;10:e0124165.
Traverso A, Wee L, Dekker A, Gillies R. Repeatability and reproducibility of radiomic features: a systematic review. Int J Radiat Oncol Biol Phys 2018;102:1143–1158.
Balachandran VP, Gonen M, Smith JJ, DeMatteo RP. Nomograms in oncology: more than meets the eye. Lancet Oncol 2015;16:e173–e180.
Mazurowski MA. Radiogenomics: what it is and why it is important. J Am Coll Radiol 2015;12:862–866.
Acknowledgments
The authors thank all colleagues who participated in collecting and organizing the data.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 81874058)
Author information
Authors and Affiliations
Contributions
NL collected and organized the clinical data. XG completed the modeling and data analysis and wrote the manuscript. JPZ directed the research. All authors contributed to the article and approved the submitted version.
Corresponding author
Ethics declarations
Conflict of interest
All authors have no conflicts of interest to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc/4.0/.
About this article
Cite this article
Guan, X., Lu, N. & Zhang, J. Computed Tomography-Based Deep Learning Nomogram Can Accurately Predict Lymph Node Metastasis in Gastric Cancer. Dig Dis Sci 68, 1473–1481 (2023). https://doi.org/10.1007/s10620-022-07640-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10620-022-07640-3