Abstract
Approximately 80% of the human genome harbors biochemical marks of active transcription that its majority transcribes to noncoding RNAs, namely long noncoding RNAs (lncRNAs). LncRNAs are heterogeneous RNA transcripts that regulate critical biological processes such as cell survival and death. They involve in the progression of different cancers by affecting transcriptional and post-transcriptional modifications as well as epigenetic control of numerous tumor suppressors and oncogenes. Recent findings show that aberrant expression of lncRNAs is associated with tumor initiation, progression, invasion, and overall survival of patients with gastrointestinal (GI) cancers. Some lncRNAs play as tumor suppressors in all GI cancers, but others play as tumor promoters. However, some other lncRNAs might function as a tumor suppressor in one GI cancer, but as a tumor promoter in another GI cancer type. This fact highlights possible context dependency of the expression patterns and roles of at least some lncRNAs in GI cancer development and progression. Here, we review the functional relation of lncRNAs involved in the development and progression of GI cancer by focusing on their roles as tumor suppressor and tumor promoter genes.



Similar content being viewed by others
References
Xie H, Ma B, Gao Q, Zhan H, et al. Long non-coding RNA CRNDE in cancer prognosis: review and meta-analysis. Clinica Chimica Acta. 2018;485:262–271.
Aguadé-Gorgorió G, Solé R. Adaptive dynamics of unstable cancer populations: the canonical equation. Evol Appl. 2018;11:1283–1292.
Wu K, Zhang N, Ma J, Huang J, et al. Long noncoding RNA FAL1 promotes proliferation and inhibits apoptosis of human colon cancer cells. IUBMB Life. 2018;70:1093–1100.
Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12.
Lee EY, Muller WJ. Oncogenes and tumor suppressor genes. Cold Spring Harbor Perspect Biol. 2010;2:a003236.
Baeissa HM, Benstead-Hume G, Richardson CJ, Pearl FM. Mutational patterns in oncogenes and tumour suppressors. Biochem Soc Trans. 2016;44:925–931.
Djebali S, Davis CA, Merkel A, Dobin A, et al. Landscape of transcription in human cells. Nature. 2012;489:101.
Consortium F. A promoter-level mammalian expression atlas. Nature. 2014;507:462.
Balas MM, Johnson AM. Exploring the mechanisms behind long noncoding RNAs and cancer. Non-Coding RNA Res. 2018;3:108–117.
Sun W, Yang Y, Xu C, Xie Y, Guo J. Roles of long noncoding RNAs in gastric cancer and their clinical applications. J Cancer Res Clin Oncol. 2016;142:2231–2237.
Lu S, Su Z, Fu W, Cui Z, et al. Altered expression of long non-coding RNA GAS5 in digestive tumors. Biosci Rep. 2019;39:BSR20180789.
Zeng S, Xiao Y-F, Tang B, Hu C-J, et al. Long noncoding RNA in digestive tract cancers: function, mechanism, and potential biomarker. The Oncologist. 2015;20:898–906.
Parsons C, Tayoun AM, Benando B, Ragusa G, et al. The role of long noncoding RNAs in cancer metastasis. J Cancer Metastasis Treat. 2018;4:1–23.
Fanelli GN, Gasparini P, Coati I, Cui R, et al. LONG-NONCODING RNAs in gastroesophageal cancers. Non-coding RNA Res. 2018;3:195–212.
Zheng Y, Yang C, Tong S, Ding Y, et al. Genetic variation of long non-coding RNA TINCR contribute to the susceptibility and progression of colorectal cancer. Oncotarget. 2017;8:33536.
Ding Z, Lan H, Xu R, Zhou X, Pan Y. LncRNA TP73-AS1 accelerates tumor progression in gastric cancer through regulating miR-194-5p/SDAD1 axis. Pathol-Res Pract. 2018;214:1993–1999.
Zhan L, Li J, Wei B. Long non-coding RNAs in ovarian cancer. J Exp Clin Cancer Res. 2018;37:120.
Luo Y, Ouyang J, Zhou D, Zhong S, et al. Long noncoding RNA GAPLINC promotes cells migration and invasion in colorectal cancer cell by regulating miR-34a/c-MET signal pathway. Digest Dis Sci. 2018;63:890–899.
Chen S, Zhu J, Wang F, Guan Z, et al. LncRNAs and their role in cancer stem cells. Oncotarget. 2017;8:110685.
Sanchez Calle A, Kawamura Y, Yamamoto Y, Takeshita F, Ochiya T. Emerging roles of long non-coding RNA in cancer. Cancer Sci. 2018;109:2093–2100.
Jin L, He Y, Tang S, Huang S. LncRNA GHET1 predicts poor prognosis in hepatocellular carcinoma and promotes cell proliferation by silencing KLF2. J Cell Physiol. 2018;233:4726–4734.
Adriaens C, Marine J-C. NEAT1-containing Paraspeckles: central hubs in stress response and tumor formation. Cell Cycle. 2017;16:137.
Ma D, Cao Y, Wang Z, He J, et al. CCAT1 lncRNA promotes inflammatory bowel disease malignancy by destroying intestinal barrier via downregulating miR-185-3p. Inflam Bowel Dis. 2019;25:862–874.
Huang T, Wang M, Huang B, Chang A, et al. Long noncoding RNAs in the mTOR signaling network: biomarkers and therapeutic targets. Apoptosis. 2018;23:255–264.
Ding J, Li D, Gong M, Wang J, et al. Expression and clinical significance of the long non-coding RNA PVT1 in human gastric cancer. OncoTargets Ther. 2014;7:1625.
Zhang X-W, Bu P, Liu L, Zhang X-Z, Li J. Overexpression of long non-coding RNA PVT1 in gastric cancer cells promotes the development of multidrug resistance. Biochem Biophys Res Commun. 2015;462:227–232.
Takahashi Y, Sawada G, Kurashige J, Uchi R, et al. Amplification of PVT-1 is involved in poor prognosis via apoptosis inhibition in colorectal cancers. Br J Cancer. 2014;110:164.
Guo J, Hao C, Wang C, Li L. Long noncoding RNA PVT1 modulates hepatocellular carcinoma cell proliferation and apoptosis by recruiting EZH2. Cancer Cell Int. 2018;18:98.
Wu B-Q, Jiang Y, Zhu F, Sun D-L, He X-Z. Long noncoding RNA PVT1 promotes EMT and cell proliferation and migration through downregulating p21 in pancreatic cancer cells. Technol Cancer Res Treatment. 2017;16:819–827.
Li P-D, Hu J-L, Ma C, Ma H, et al. Upregulation of the long non-coding RNA PVT1 promotes esophageal squamous cell carcinoma progression by acting as a molecular sponge of miR-203 and LASP1. Oncotarget. 2017;8:34164.
Chen J, Yu Y, Li H, Hu Q, et al. Long non-coding RNA PVT1 promotes tumor progression by regulating the miR-143/HK2 axis in gallbladder cancer. Molecular Cancer. 2019;18:33.
Zhao J, Du P, Cui P, Qin Y, et al. LncRNA PVT1 promotes angiogenesis via activating the STAT3/VEGFA axis in gastric cancer. Oncogene. 2018;37:4094.
Xu M-D, Wang Y, Weng W, Wei P, et al. A positive feedback loop of lncRNA-PVT1 and FOXM1 facilitates gastric cancer growth and invasion. Clin Cancer Res. 2017;23:2071–2080.
Kong R, Zhang E-B, Yin D-D, You L-H, et al. Long noncoding RNA PVT1 indicates a poor prognosis of gastric cancer and promotes cell proliferation through epigenetically regulating p15 and p16. Mol Cancer. 2015;14:82.
Huppi K, Volfovsky N, Runfola T, Jones TL, et al. The identification of microRNAs in a genomically unstable region of human chromosome 8q24. Mol Cancer Res. 2008;6:212–221.
Li T, Meng X-L, Yang W-Q. Long noncoding RNA PVT1 acts as a “Sponge” to inhibit microRNA-152 in gastric cancer cells. Digest Dis Sci. 2017;62:3021–3028.
Huang T, Liu HW, Chen JQ, Wang SH, et al. The long noncoding RNA PVT1 functions as a competing endogenous RNA by sponging miR-186 in gastric cancer. Biomed Pharmacother. 2017;88:302–308.
Xin Y, Li Z, Shen J, Chan MT, Wu WKK. CCAT 1: a pivotal oncogenic long non-coding RNA in human cancers. Cell Prolifer. 2016;49:255–260.
Mizrahi I, Mazeh H, Grinbaum R, Beglaibter N, et al. Colon cancer associated transcript-1 (CCAT1) expression in adenocarcinoma of the stomach. J Cancer. 2015;6:105.
Li Y, Jing F, Ding Y, He Q, et al. Long noncoding RNA CCAT1 polymorphisms are associated with the risk of colorectal cancer. Cancer Genet. 2018;222:13–19.
Yang F, Xue X, Bi J, Zheng L, et al. Long noncoding RNA CCAT1, which could be activated by c-Myc, promotes the progression of gastric carcinoma. J Cancer Res Clin Oncol. 2013;139:437–445.
He X, Tan X, Wang X, Jin H, et al. C-Myc-activated long noncoding RNA CCAT1 promotes colon cancer cell proliferation and invasion. Tumor Biol. 2014;35:12181–12188.
Li B, Shi C, Zhao J, Li B. Long noncoding RNA CCAT1 functions as a ceRNA to antagonize the effect of miR-410 on the down-regulation of ITPKB in human HCT-116 and HCT-8 cells. Oncotarget. 2017;8:92855.
Li Y, Zhu G, Ma Y, Qu H. LncRNA CCAT1 contributes to the growth and invasion of gastric cancer via targeting miR-219-1. J Cell Biochem. 2017.
Zhou B, Wang Y, Jiang J, Jiang H, et al. The long noncoding RNA colon cancer-associated transcript-1/miR-490 axis regulates gastric cancer cell migration by targeting hnRNPA1. IUBMB Life. 2016;68:201–210.
Zhu H, Zhao H, Zhang L, Xu J, et al. Dandelion root extract suppressed gastric cancer cells proliferation and migration through targeting lncRNA-CCAT1. Biomed Pharmacother. 2017;93:1010–1017.
Zhu H-Q, Zhou X, Chang H, Li H-G, et al. Aberrant expression of CCAT1 regulated by c-Myc predicts the prognosis of hepatocellular carcinoma. Asian Pac J Cancer Prev. 2015;16:5181–5185.
Deng L, Yang S-B, Xu F-F, Zhang J-H. Long noncoding RNA CCAT1 promotes hepatocellular carcinoma progression by functioning as let-7 sponge. J Exp Clin Cancer Res. 2015;34:18.
Yu Q, Zhou X, Xia Q, Shen J, et al. Long non-coding RNA CCAT1 that can be activated by c-Myc promotes pancreatic cancer cell proliferation and migration. Am J Transl Res. 2016;8:5444.
Zhang E, Han L, Yin D, He X, et al. H3K27 acetylation activated-long non-coding RNA CCAT1 affects cell proliferation and migration by regulating SPRY4 and HOXB13 expression in esophageal squamous cell carcinoma. Nucl Acids Res. 2016;45:3086–3101.
Hu Y, Wang J, Qian J, Kong X, et al. Long noncoding RNA GAPLINC regulates CD44-dependent cell invasiveness and associates with poor prognosis of gastric cancer. Cancer Res. 2014;74:6890–6902.
Yang P, Chen T, Xu Z, Zhu H, et al. Long noncoding RNA GAPLINC promotes invasion in colorectal cancer by targeting SNAI2 through binding with PSF and NONO. Oncotarget. 2016;7:42183.
Diao L, Wang S, Sun Z. Long noncoding RNA GAPLINC promotes gastric cancer cell proliferation by acting as a molecular sponge of miR-378 to modulate MAPK1 expression. OncoTargets Ther. 2018;11:2797.
Liu L, Zhao X, Zou H, Bai R, et al. Hypoxia promotes gastric cancer malignancy partly through the HIF-1α dependent transcriptional activation of the long non-coding RNA GAPLINC. Front Physiol. 2016;7:420.
Liu J, Liu L, Wan J-X, Song Y. Long noncoding RNA SNHG20 promotes gastric cancer progression by inhibiting p21 expression and regulating the GSK-3β/β-catenin signaling pathway. Oncotarget. 2017;8:80700.
Wu J, Zhao W, Wang Z, Xiang X, et al. Long non-coding RNA SNHG20 promotes the tumorigenesis of oral squamous cell carcinoma via targeting miR-197/LIN28 axis. J Cell Mol Med. 2019;23:680–688.
Li C, Zhou L, He J, Fang X-Q, et al. Increased long noncoding RNA SNHG20 predicts poor prognosis in colorectal cancer. BMC Cancer. 2016;16:655.
Cui N, Liu J, Xia H, Xu D. LncRNA SNHG20 contributes to cell proliferation and invasion by upregulating ZFX expression sponging miR-495-3p in gastric cancer. J Cell Biochem. 2019;120:3114–3123.
Gao P, Fan R, Ge T. SNHG20 serves as a predictor for prognosis and promotes cell growth in oral squamous cell carcinoma. Oncol Lett. 2019;17:951–957.
Zhang D, Cao C, Liu L, Wu D. Up-regulation of LncRNA SNHG20 predicts poor prognosis in hepatocellular carcinoma. J Cancer. 2016;7:608.
Liu J, Lu C, Xiao M, Jiang F, et al. Long non-coding RNA SNHG20 predicts a poor prognosis for HCC and promotes cell invasion by regulating the epithelial-to-mesenchymal transition. Biomed Pharmacother. 2017;89:857–863.
Zhang C, Jiang F, Su C, Xie P, Xu L. Upregulation of long noncoding RNA SNHG20 promotes cell growth and metastasis in esophageal squamous cell carcinoma via modulating ATM-JAK-PD-L1 pathway. J Cell Biochem. 2019;120:11642–11650.
Wang L, Jiang F, Xia X, Zhang B. LncRNA FAL1 promotes carcinogenesis by regulation of miR-637/NUPR1 pathway in colorectal cancer. Int J Biochem Cell Biol. 2019;106:46–56.
Li B, Mao R, Liu C, Zhang W, et al. LncRNA FAL1 promotes cell proliferation and migration by acting as a CeRNA of miR-1236 in hepatocellular carcinoma cells. Life Sci. 2018;197:122–129.
Liu T, Wang Z, Zhou R, Liang W. Focally amplified lncRNA on chromosome 1 regulates apoptosis of esophageal cancer cells via DRP1 and mitochondrial dynamics. IUBMB Life. 2019;71:254–260.
Zhu C, Xiao D, Dai L, Xu H, et al. Highly expressed lncRNA FAL1 promotes the progression of gastric cancer by inhibiting PTEN. Eur Rev Med Pharmacol Sci. 2018;22:8257–8264.
Yang F, Xue X, Zheng L, Bi J, et al. Long non-coding RNA GHET1 promotes gastric carcinoma cell proliferation by increasing c-Myc mRNA stability. FEBS J. 2014;281:802–813.
Xia Y, Yan Z, Wan Y, Wei S, et al. Knockdown of long noncoding RNA GHET1 inhibits cell-cycle progression and invasion of gastric cancer cells. Mol Med Rep. 2018;18:3375–3381.
Liu H, Zhen Q, Fan Y. LncRNA GHET1 promotes esophageal squamous cell carcinoma cells proliferation and invasion via induction of EMT. Int J Biol Mark. 2017;32:403–408.
Zhou J, Li X, Wu M, Lin C, et al. Knockdown of long noncoding RNA GHET1 inhibits cell proliferation and invasion of colorectal cancer. Oncol Res Featur Preclin Clin Cancer Therap. 2016;23:303–309.
Li J, Jiang X, Li Z, Huang L, et al. Long noncoding RNA GHET1 in human cancer. Clinica Chimica Acta. 2018.
Huang H, Liao W, Zhu X, Liu H, Cai L. Knockdown of long noncoding RNA GHET1 inhibits cell activation of gastric cancer. Biomed Pharmacother. 2017;92:562–568.
Ding G, Li W, Liu J, Zeng Y, et al. LncRNA GHET1 activated by H3K27 acetylation promotes cell tumorigenesis through regulating ATF1 in hepatocellular carcinoma. Biomed Pharmacother. 2017;94:326–331.
Wei C-C, Nie F-Q, Jiang L-L, Chen Q-N, et al. The pseudogene DUXAP10 promotes an aggressive phenotype through binding with LSD1 and repressing LATS2 and RRAD in non small cell lung cancer. Oncotarget. 2017;8:5233.
Wang Z, Ren B, Huang J, Yin R, et al. LncRNA DUXAP10 modulates cell proliferation in esophageal squamous cell carcinoma through epigenetically silencing p21. Cancer Biol Ther. 2018;19:998–1005.
Lian Y, Xiao C, Yan C, Chen D, et al. Knockdown of pseudogene derived from lncRNA DUXAP10 inhibits cell proliferation, migration, invasion, and promotes apoptosis in pancreatic cancer. J Cell Biochem. 2018;119:3671–3682.
Xu Y, Yu X, Wei C, Nie F, et al. Over-expression of oncigenic pesudogene DUXAP10 promotes cell proliferation and invasion by regulating LATS1 and β-catenin in gastric cancer. J Exp Clin Cancer Res. 2018;37:13.
Lian Y, Xu Y, Xiao C, Xia R, et al. The pseudogene derived from long non-coding RNA DUXAP10 promotes colorectal cancer cell growth through epigenetically silencing of p21 and PTEN. Sci Rep. 2017;7:7312.
Shang C, Sun L, Zhang J, Zhao B, et al. Silence of cancer susceptibility candidate 9 inhibits gastric cancer and reverses chemoresistance. Oncotarget. 2017;8:15393.
Wu Y, Hu L, Liang Y, Li J, et al. Up-regulation of lncRNA CASC9 promotes esophageal squamous cell carcinoma growth by negatively regulating PDCD4 expression through EZH2. Mol Cancer. 2017;16:150.
Klingenberg M, Groß M, Goyal A, Polycarpou-Schwarz M, et al. The lncRNA CASC9 and RNA binding protein HNRNPL form a complex and co-regulate genes linked to AKT signaling. Hepatology. 2018;68:1817–1832.
Pan Z, Mao W, Bao Y, Zhang M, et al. The long noncoding RNA CASC9 regulates migration and invasion in esophageal cancer. Cancer Med. 2016;5:2442–2447.
Liang Y, Chen X, Wu Y, Li J, et al. LncRNA CASC9 promotes esophageal squamous cell carcinoma metastasis through upregulating LAMC2 expression by interacting with the CREB-binding protein. Cell Death Differ. 2018;25:1980.
Leng C, Zhang Z-G, Chen W-X, Luo H-P, et al. An integrin beta4-EGFR unit promotes hepatocellular carcinoma lung metastases by enhancing anchorage independence through activation of FAK–AKT pathway. Cancer Lett. 2016;376:188–196.
Fei T, Chen Y, Xiao T, Li W, et al. Genome-wide CRISPR screen identifies HNRNPL as a prostate cancer dependency regulating RNA splicing. Pro Natl Acad Sci. 2017;114:E5207–E5215.
Yang Y, Chen D, Liu H, Yang K. Increased expression of lncRNA CASC9 promotes tumor progression by suppressing autophagy-mediated cell apoptosis via the AKT/mTOR pathway in oral squamous cell carcinoma. Cell Death Dis. 2019;10:41.
Chen G, Sun W, Hua X, Zeng W, Yang L. Long non-coding RNA FOXD2-AS1 aggravates nasopharyngeal carcinoma carcinogenesis by modulating miR-363-5p/S100A1 pathway. Gene. 2018;645:76–84.
Yang X, Duan B, Zhou X. Long non-coding RNA FOXD2-AS1 functions as a tumor promoter in colorectal cancer by regulating EMT and Notch signaling pathway. Eur Rev Med Pharmacol Sci. 2017;21:3586–3591.
Chang Y, Zhang J, Zhou C, Qiu G, et al. Long non-coding RNA FOXD2-AS1 plays an oncogenic role in hepatocellular carcinoma by targeting miR-206. Oncol Rep. 2018;40:3625–3634.
Bao J, Zhou C, Zhang J, Mo J, et al. Upregulation of the long noncoding RNA FOXD2-AS1 predicts poor prognosis in esophageal squamous cell carcinoma. Cancer Biom. 2018;21:527–533.
Xu T-P, Wang W-Y, Ma P, Shuai Y, et al. Upregulation of the long noncoding RNA FOXD2-AS1 promotes carcinogenesis by epigenetically silencing EphB3 through EZH2 and LSD1, and predicts poor prognosis in gastric cancer. Oncogene. 2018;37:5020–5036.
Zhu Y, Qiao L, Zhou Y, Ma N, et al. Long non-coding RNA FOXD 2-AS 1 contributes to colorectal cancer proliferation through its interaction with micro RNA-185-5p. Cancer Sci. 2018;109:2235–2242.
Yan Y, Li S, Wang S, Rubegni P, et al. Long noncoding RNA HAND2-AS1 inhibits cancer cell proliferation, migration, and invasion in esophagus squamous cell carcinoma by regulating microRNA-21. J Cell Biochem. 2019;120:9564–9571.
Shi B, Zhang X, Chao L, Zheng Y, et al. Comprehensive analysis of key genes, microRNAs and long non-coding RNAs in hepatocellular carcinoma. FEBS Open Biol. 2018;8:1424–1436.
Zhou J, Lin J, Zhang H, Zhu F, Xie R. LncRNA HAND2-AS1 sponging miR-1275 suppresses colorectal cancer progression by upregulating KLF14. Biochem Biophys Res Commun. 2018;503:1848–1853.
Wan L, Kong J, Tang J, Wu Y, et al. HOTAIRM 1 as a potential biomarker for diagnosis of colorectal cancer functions the role in the tumour suppressor. J Cell Mol Med. 2016;20:2036–2044.
Lu R, Zhao G, Yang Y, Jiang Z, et al. Long noncoding RNA HOTAIRM1 inhibits cell progression by regulating miR-17-5p/PTEN axis in gastric cancer. J Cell Biochem. 2019;120:4952–4965.
Zhou Y, Gong B, Jiang Z-L, Zhong S, et al. Microarray expression profile analysis of long non-coding RNAs in pancreatic ductal adenocarcinoma. Int J Oncol. 2016;48:670–680.
Zhang Y, Mi L, Xuan Y, Gao C, et al. LncRNA HOTAIRM1 inhibits the progression of hepatocellular carcinoma by inhibiting the Wnt signaling pathway. Eur Rev Med Pharmacol Sci. 2018;22:4861–4868.
Liu L, Zhang D, Wu D. 748 long non-coding RNA miR22HG represses hepatocellular carcinoma cell invasion by deriving miR-22 and targeting HMGB1. Gastroenterology. 2016;150:S1045–S1046.
Dong Y, Yan W, Zhang S-L, Zhang M-Z-H, et al. Prognostic values of long non-coding RNA MIR22HG for patients with hepatocellular carcinoma after hepatectomy. Oncotarget. 2017;8:114041.
Wu Y, Wang PS, Wang BG, Xu L, et al. Genomewide identification of a novel six-LncRNA signature to improve prognosis prediction in resectable hepatocellular carcinoma. Cancer Med. 2018;7:6219–6233.
Cui Z, An X, Li J, Liu Q, Liu W. LncRNA MIR22HG negatively regulates miR-141-3p to enhance DAPK1 expression and inhibits endometrial carcinoma cells proliferation. Biomed Pharmacother. 2018;104:223–228.
Su W, Feng S, Chen X, Yang X, et al. Silencing of long noncoding RNA MIR22HG triggers cell survival/death signaling via oncogenes YBX1, MET, and p21 in lung cancer. Cancer Res. 2018;78:3207–3219.
Zhang D-Y, Zou X-J, Cao C-H, Zhang T, et al. Identification and functional characterization of long non-coding RNA MIR22HG as a tumor suppressor for hepatocellular carcinoma. Theranostics. 2018;8:3751.
Dai W, Zhang G, Makeyev EV. RNA-binding protein HuR autoregulates its expression by promoting alternative polyadenylation site usage. Nucl Acids Res. 2011;40:787–800.
Wu Y, Zhou Y, Huan L, Xu L, et al. LncRNA MIR22HG inhibits growth, migration and invasion through regulating the miR-10a-5p/NCOR2 axis in hepatocellular carcinoma cells. Cancer Sci. 2019;110:973.
Li H, Wang Y. Long noncoding RNA (lncRNA) MIR22HG suppresses gastric cancer progression through attenuating NOTCH2 signaling. Med Sci Monitor Int Med J Exp Clin Res. 2019;25:656.
Song H, Sun W, Ye G, Ding X, et al. Long non-coding RNA expression profile in human gastric cancer and its clinical significances. J Transl Med. 2013;11:225.
Yue B, Sun B, Liu C, Zhao S, et al. Long non-coding RNA Fer-1-like protein 4 suppresses oncogenesis and exhibits prognostic value by associating with miR-106a-5p in colon cancer. Cancer Sci. 2015;106:1323–1332.
Ma W, Zhang C, Li H, Gu J, et al. LncRNA FER1L4 suppressed cancer cell growth and invasion in esophageal squamous cell carcinoma. Eur Rev Med Pharmacol Sci. 2018;22:2638–2645.
Xia T, Chen S, Jiang Z, Shao Y, et al. Long noncoding RNA FER1L4 suppresses cancer cell growth by acting as a competing endogenous RNA and regulating PTEN expression. Sci Rep. 2015;5:13445.
Wu J, Huang J, Wang W, Xu J, et al. Long non-coding RNA Fer-1-like protein 4 acts as a tumor suppressor via miR-106a-5p and predicts good prognosis in hepatocellular carcinoma. Cancer Biom. 2017;20:55–65.
Xia T, Liao Q, Jiang X, Shao Y, et al. Long noncoding RNA associated-competing endogenous RNAs in gastric cancer. Sci Rep. 2014;4:6088.
Sun X, Zheng G, Li C, Liu C. Long non-coding RNA Fer-1-like family member 4 suppresses hepatocellular carcinoma cell proliferation by regulating PTEN in vitro and in vivo. Mol Med Rep. 2019;19:685–692.
Wang PL, Liu B, Xia Y, Pan CF, et al. Long non-coding RNA-Low Expression in Tumor inhibits the invasion and metastasis of esophageal squamous cell carcinoma by regulating p53 expression. Mol Med Rep. 2016;13:3074–3082.
Weidle UH, Birzele F, Kollmorgen G, Rueger R. Long non-coding RNAs and their role in metastasis. Cancer Genom-Proteom. 2017;14:143–160.
Chen Z, Lin J, Wu S, Xu C, et al. Up-regulated miR-548 k promotes esophageal squamous cell carcinoma progression via targeting long noncoding RNA-LET. Exp Cell Res. 2018;362:90–101.
Tian J, Hu X, Gao W, Zhang J, et al. Identification of the long non-coding RNA LET as a novel tumor suppressor in gastric cancer. Mol Med Rep. 2017;15:2229–2234.
Ma MZ, Kong X, Weng MZ, Zhang MD, et al. Long non-coding RNA-LET is a positive prognostic factor and exhibits tumor-suppressive activity in gallbladder cancer. Mol Carcinogen. 2015;54:1397–1406.
Mao Z, Li H, Du B, Cui K, et al. LncRNA DANCR promotes migration and invasion through suppression of lncRNA-LET in gastric cancer cells. Biosci Rep. 2017;37:BSR20171070.
Zhou B, Jing X-Y, Wu J-Q, Xi H-F, Lu G-J. Down-regulation of long non-coding RNA LET is associated with poor prognosis in gastric cancer. Int J Clin Exp Pathol. 2014;7:8893.
He Y, Meng X-M, Huang C, Wu B-M, et al. Long noncoding RNAs: novel insights into hepatocelluar carcinoma. Cancer Lett. 2014;344:20–27.
Xu T, Liu X, Xia R, Yin L, et al. SP1-induced upregulation of the long noncoding RNA TINCR regulates cell proliferation and apoptosis by affecting KLF2 mRNA stability in gastric cancer. Oncogene. 2015;34:5648.
Xu Y, Qiu M, Chen Y, Wang J, et al. Long noncoding RNA, tissue differentiation-inducing nonprotein coding RNA is upregulated and promotes development of esophageal squamous cell carcinoma. Dis Esophagus. 2016;29:950–958.
Tian F, Xu J, Xue F, Guan E, Xu X. TINCR expression is associated with unfavorable prognosis in patients with hepatocellular carcinoma. Biosci Rep. 2017;37:301.
Xu T-P, Wang Y-F, Xiong W-L, Ma P, et al. E2F1 induces TINCR transcriptional activity and accelerates gastric cancer progression via activation of TINCR/STAU1/CDKN2B signaling axis. Cell Death Dis. 2017;8:e2837.
Chen Z, Liu H, Yang H, Gao Y, et al. The long noncoding RNA, TINCR, functions as a competing endogenous RNA to regulate PDK1 expression by sponging miR-375 in gastric cancer. OncoTargets Ther. 2017;10:3353.
Zhang Z-y, Lu Y-x, Zhang Z-y, Chang Y-y, et al. Loss of TINCR expression promotes proliferation, metastasis through activating EpCAM cleavage in colorectal cancer. Oncotarget. 2016;7:22639.
Zhang X, Yao J, Shi H, Gao B, Zhang L. LncRNA TINCR/microRNA-107/CD36 regulates cell proliferation and apoptosis in colorectal cancer via PPAR signaling pathway based on bioinformatics analysis. Biol Chem. 2019;400:663–675.
Pang JCS, Li KKW, Lau KM, Ng YL, et al. KIAA0495/PDAM is frequently downregulated in oligodendroglial tumors and its knockdown by siRNA induces cisplatin resistance in glioma cells. Brain Pathol. 2010;20:1021–1032.
Jia Z, Peng J, Yang Z, Chen J, et al. Long non-coding RNA TP73-AS1 promotes colorectal cancer proliferation by acting as a ceRNA for miR-103 to regulate PTEN expression. Gene. 2019;685:222–229.
Cai Y, Yan P, Zhang G, Yang W, et al. Long non-coding RNA TP73-AS1 sponges miR-194 to promote colorectal cancer cell proliferation, migration and invasion via up-regulating TGFα. Cancer Biom. 2018(Preprint):1–12.
Li S, Huang Y, Huang Y, Fu Y, et al. The long non-coding RNA TP73-AS1 modulates HCC cell proliferation through miR-200a-dependent HMGB1/RAGE regulation. J Exp Clin Cancer Res. 2017;36:51.
Peng J. si-TP73-AS1 suppressed proliferation and increased the chemotherapeutic response of GC cells to cisplatin. Oncol Lett. 2018;16:3706–3714.
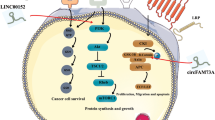
Xiong X, Shi Q, Yang X, Wang W, Tao J. LINC00052 functions as a tumor suppressor through negatively modulating miR-330-3p in pancreatic cancer. J Cell Physiol. 2019;234:15619–15626.
Xiong D, Sheng Y, Ding S, Chen J, et al. LINC00052 regulates the expression of NTRK3 by miR-128 and miR-485-3p to strengthen HCC cells invasion and migration. Oncotarget. 2016;7:47593.
Zhu L, Yang N, Chen J, Zeng T, et al. LINC00052 upregulates EPB41L3 to inhibit migration and invasion of hepatocellular carcinoma by binding miR-452-5p. Oncotarget. 2017;8:63724.
Yan S, Shan X, Chen K, Liu Y, et al. LINC00052/miR-101-3p axis inhibits cell proliferation and metastasis by targeting SOX9 in hepatocellular carcinoma. Gene. 2018;679:138–149.
Shan Y, Ying R, Jia Z, Kong W, et al. LINC00052 promotes gastric cancer cell proliferation and metastasis via activating the Wnt/β-catenin signaling pathway. Oncol Res Featur Preclin Clin Cancer Therap. 2017;25:1589–1599.
Gold JM, Raja A. Cisplatin (Cisplatinum). StatPearls [Internet]: StatPearls Publishing; 2019.
You L, Chang D, Du H-Z, Zhao Y-P. Genome-wide screen identifies PVT1 as a regulator of Gemcitabine sensitivity in human pancreatic cancer cells. Biochem Biophys Res Commun. 2011;407:1–6.
Hu M, Zhang Q, Tian XH, Wang JL, et al. lncRNA CCAT1 is a biomarker for the proliferation and drug resistance of esophageal cancer via the miR-143/PLK1/BUBR1 axis. Mol Carcinogen. 2019;58:2207–2217.
McCleland ML, Mesh K, Lorenzana E, Chopra VS, et al. CCAT1 is an enhancer-templated RNA that predicts BET sensitivity in colorectal cancer. J Clin Investig. 2016;126:639–652.
Yu J, Shen J, Qiao X, Cao L, et al. SNHG20/miR-140-5p/NDRG3 axis contributes to 5-fluorouracil resistance in gastric cancer. Oncol Lett. 2019;18:1337–1343.
Zhang N, Yin Y, Xu S-J, Chen W-S. 5-Fluorouracil: mechanisms of resistance and reversal strategies. Molecules. 2008;13:1551–1569.
Wang W, Li Y, Li Y, Hong A, et al. NDRG3 is an androgen regulated and prostate enriched gene that promotes in vitro and in vivo prostate cancer cell growth. Int J Cancer. 2009;124:521–530.
Jing J-S, Li H, Wang S-C, Ma J-M, et al. NDRG3 overexpression is associated with a poor prognosis in patients with hepatocellular carcinoma. Biosci Rep. 2018;38:1–9.
Verstraelen J, Reichl S. Multidrug resistance-associated protein (MRP1, 2, 4 and 5) expression in human corneal cell culture models and animal corneal tissue. Mol Pharm. 2014;11:2160–2171.
Zhang X, Bo P, Liu L, Zhang X, Li J. Overexpression of long non-coding RNA GHET1 promotes the development of multidrug resistance in gastric cancer cells. Biomed Pharmacother. 2017;92:580–585.
Weaver BA. How Taxol/paclitaxel kills cancer cells. Mol Biol Cell. 2014;25:2677–2681.
Thorn CF, Oshiro C, Marsh S, Hernandez-Boussard T, et al. Doxorubicin pathways: pharmacodynamics and adverse effects. Pharmacogenet Genom. 2011;21:440.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Khajehdehi, M., Khalaj-Kondori, M., Ghasemi, T. et al. Long Noncoding RNAs in Gastrointestinal Cancer: Tumor Suppression Versus Tumor Promotion. Dig Dis Sci 66, 381–397 (2021). https://doi.org/10.1007/s10620-020-06200-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10620-020-06200-x