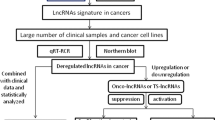
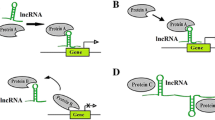
Abstract
A complex sequence of occurrences, including host genetic vulnerability, Helicobacter pylori infection, and other environmental variables, culminate in gastric cancer (GC). The development of several genetic and epigenetic changes in oncogenes and tumor suppressor genes causes dysregulation of several signaling pathways, which upsets the cell cycle and the equilibrium between cell division and apoptosis, leading to GC. Developments in computational biology and RNA-seq technology enable quick detection and characterization of long non-coding RNAs (lncRNAs). Recent studies have shown that long non-coding RNAs (lncRNAs) have multiple roles in the development of gastric cancer. These lncRNAs interact with molecules of protein, RNA, DNA, and/or combinations. This review article explores several gastric cancer-associated lncRNAs, such as ADAMTS9-AS2, UCA1, XBP-1, and LINC00152. These various lncRNAs could change GC cell apoptosis, migration, and invasion features in the tumor microenvironment. This review provides an overview of the most recent research on lncRNAs and GC cell apoptosis, migration, invasion, and drug resistance, focusing on studies conducted in cancer cells and healthy cells during differentiation.

Similar content being viewed by others
Data availability
No datasets were generated or analysed during the current study.
References
Zou Y, Chen B (2021) Long non-coding RNA HCP5 in cancer. Clinica Chimica Acta. Int J Clin Chem 512:33–39
Zakutansky PM, Feng Y (2022) The long non-coding RNA GOMAFU in Schizophrenia: function, Disease Risk, and Beyond. Cells. ;11
Yan H, Bu P (2021) Non-coding RNA in cancer. Essays Biochem 65:625–639
Sharma U, Barwal TS, Malhotra A, Pant N, Vivek, Dey D et al (2020) Long non-coding RNA TINCR as potential biomarker and therapeutic target for cancer. Life Sci 257:118035
Qian Y, Shi L, Luo Z (2020) Long non-coding RNAs in Cancer: implications for diagnosis, prognosis, and Therapy. Front Med 7:612393
Kanwal F, Lu C (2019) A review on native and denaturing purification methods for non-coding RNA (ncRNA). J Chromatogr B 1120:71–79
Chen B, Dragomir MP, Yang C, Li Q, Horst D, Calin GA (2022) Targeting non-coding RNAs to overcome cancer therapy resistance. Signal Transduct Target Therapy 7:121
Entezari M, Sadrkhanloo M, Rashidi M, Asnaf SE, Taheriazam A, Hashemi M et al (2022) Non-coding RNAs and macrophage interaction in tumor progression. Crit Rev Oncol/Hematol 173:103680
Jim MA, Pinheiro PS, Carreira H, Espey DK, Wiggins CL, Weir HK (2017) Stomach cancer survival in the United States by race and stage (2001-2009): findings from the CONCORD‐2 study. Cancer 123:4994–5013
Li X, Wu Z, Fu X, Han W (2014) lncRNAs: insights into their function and mechanics in underlying disorders. Mutat Research/Reviews Mutat Res 762:1–21
Evans JR, Feng FY, Chinnaiyan AM (2016) The bright side of dark matter: lncRNAs in cancer. J Clin Investig 126:2775–2782
Kitagawa M, Kotake Y, Ohhata T (2012) Long non-coding RNAs involved in cancer development and cell fate determination. Curr Drug Targets 13:1616–1621
Bhat SA, Ahmad SM, Mumtaz PT, Malik AA, Dar MA, Urwat U et al (2016) Non-coding RNA Research
Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M et al (2011) Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15INK4B tumor suppressor gene. Oncogene 30:1956–1962
Congrains A, Kamide K, Ohishi M, Rakugi H (2013) ANRIL: molecular mechanisms and implications in human health. Int J Mol Sci 14:1278–1292
Berghoff EG, Clark MF, Chen S, Cajigas I, Leib DE, Kohtz JD (2013) Evf2 (Dlx6as) lncRNA regulates ultraconserved enhancer methylation and the differential transcriptional control of adjacent genes. Development 140:4407–4416
Ørom UA, Shiekhattar R (2011) Long non-coding RNAs and enhancers. Curr Opin Genet Dev 21:194–198
Bunch H, Lawney BP, Burkholder A, Ma D, Zheng X, Motola S et al (2016) RNA polymerase II promoter-proximal pausing in mammalian long non-coding genes. Genomics 108:64–77
Chen L-L (2016) Linking long non-coding RNA localization and function. Trends Biochem Sci 41:761–772
Caddle MS, Lussier RH, Heintz NH (1990) Intramolecular DNA triplexes, bent DNA and DNA unwinding elements in the initiation region of an amplified dihydrofolate reductase replicon. J Mol Biol 211:19–33
Abali EE, Skacel NE, Celikkaya H, Hsieh YC (2008) Regulation of human dihydrofolate reductase activity and expression. Vitamins Horm 79:267–292
Reece LJ, Nichols R, Ogden RC, Howell EE (1991) Construction of a synthetic gene for an R-plasmid-encoded dihydrofolate reductase and studies on the role of the N-terminus in the protein. Biochemistry 30:10895–10904
Thomson DW, Dinger ME (2016) Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet 17:272–283
Chen H, Du G, Song X, Li L (2017) Non-coding transcripts from enhancers: new insights into enhancer activity and gene expression regulation. Genom Proteom Bioinform 15:201–207
Huang S-F, Peng X-F, Jiang L, Hu CY, Ye W-C (2021) LncRNAs as therapeutic targets and potential biomarkers for lipid-related diseases. Front Pharmacol 12:729745
Pawson T, Scott JD (1997) Signaling through scaffold, anchoring, and adaptor proteins. Science 278:2075–2080
Dokland T (1999) Scaffolding proteins and their role in viral assembly. Cell Mol Life Sci CMLS 56:580–603
Kafasla P, Skliris A, Kontoyiannis DL (2014) Post-transcriptional coordination of immunological responses by RNA-binding proteins. Nat Immunol 15:492–502
Tang J, Ning R, Zeng B, Li Y (2016) Molecular evolution of PTEN pseudogenes in mammals. PLoS ONE 11:e0167851
Li W, Zhang T, Guo L, Huang L (2018) Regulation of PTEN expression by non-coding RNAs. J Experimental Clin Cancer Res 37:1–12
Wu Y, Li L, Wang Q, Zhang L, He C, Wang X et al (2020) LINC00511 promotes lung squamous cell carcinoma proliferation and migration via inhibiting mir-150-5p and activating TADA1. Translational Lung Cancer Res 9:1138
Chen J, Chen JG, Sun B, Wu J, Du C (2020) Integrative analysis of immune microenvironment-related CeRNA regulatory axis in gastric cancer. Math Biosci Eng 17:3953–3971
Foroughi K, Amini M, Atashi A, Mahmoodzadeh H, Hamann U, Manoochehri M (2018) Tissue-specific down-regulation of the long non-coding RNAs PCAT18 and LINC01133 in gastric cancer development. Int J Mol Sci 19:3881
Baldini F, Calderoni M, Vergani L, Modesto P, Florio T, Pagano A (2021) An overview of long non-coding (lnc) RNAs in neuroblastoma. Int J Mol Sci 22:4234
Mitra S (2019) Understanding the role of long non-coding RNAs in neuroblastoma development and progression
Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP (2010) A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 465:1033–1038
Yoshimoto R, Mayeda A, Yoshida M, Nakagawa S (2016) MALAT1 long non-coding RNA in cancer. Biochimica et Biophysica Acta (BBA)-Gene Regulatory mechanisms. 1859:192–199
Ouyang S, Zheng X, Zhou X, Chen Z, Yang X, Xie M (2017) LncRNA BCAR4 promotes colon cancer progression via activating Wnt/β-catenin signaling. Oncotarget 8:92815
Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ et al (2010) Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464:1071–1076
Zhou Y, Zhong Y, Wang Y, Zhang X, Batista DL, Gejman R et al (2007) Activation of p53 by MEG3 non-coding RNA. J Biol Chem 282:24731–24742
Sánchez Y, Segura V, Marín-Béjar O, Athie A, Marchese FP, González J et al (2014) Genome-wide analysis of the human p53 transcriptional network unveils a lncRNA tumour suppressor signature. Nat Commun 5:5812
Pandey GK, Mitra S, Subhash S, Hertwig F, Kanduri M, Mishra K et al (2014) The risk-associated long non-coding RNA NBAT-1 controls neuroblastoma progression by regulating cell proliferation and neuronal differentiation. Cancer Cell 26:722–737
Mourtada-Maarabouni M, Pickard M, Hedge V, Farzaneh F, Williams G (2009) GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene 28:195–208
Cavallo F, Feldman DR, Barchi M (2013) Revisiting DNA damage repair, p53-mediated apoptosis and cisplatin sensitivity in germ cell tumors. Int J Dev Biol 57:273–280
Ni K, Huang Z, Zhu Y, Xue D, Jin Q, Zhang C et al (2021) The lncRNA ADAMTS9-AS2 regulates RPL22 to modulate TNBC Progression via Controlling the TGF-β signaling pathway. Front Oncol 11:654472
Ren N, Jiang T, Wang C, Xie S, Xing Y, Piao D et al (2020) LncRNA ADAMTS9-AS2 inhibits gastric cancer (GC) development and sensitizes chemoresistant GC cells to cisplatin by regulating miR-223-3p/NLRP3 axis. Aging 12:11025
Xue M, Chen W, Li X (2016) Urothelial cancer associated 1: a long non-coding RNA with a crucial role in cancer. J Cancer Res Clin Oncol 142:1407–1419
Horita K, Kurosaki H, Nakatake M, Ito M, Kono H, Nakamura T (2019) Long non-coding RNA UCA1 enhances sensitivity to oncolytic vaccinia virus by sponging miR-18a/miR-182 and modulating the Cdc42/filopodia axis in colorectal cancer. Biochem Biophys Res Commun 516:831–838
Cheng H, Sharen G, Wang Z, Zhou J (2021) LncRNA UCA1 enhances cisplatin resistance by regulating CYP1B1-mediated apoptosis via miR-513a-3p in human gastric cancer. Cancer Manage Res. :367–377
Xu W, Wang C, Hua J (2021) X-box binding protein 1 (XBP1) function in diseases. Cell Biol Int 45:731–739
Clarke R, Cook KL, Hu R, Facey CO, Tavassoly I, Schwartz JL et al (2012) Endoplasmic reticulum stress, the unfolded protein response, autophagy, and the integrated regulation of breast cancer cell fate. Cancer Res 72:1321–1331
Zhou Y, Sha Z, Yang Y, Wu S, Chen H (2021) lncRNA NEAT1 regulates gastric carcinoma cell proliferation, invasion and apoptosis via the miR–500a–3p/XBP–1 axis. Mol Med Rep 24:1–10
Guruharsha K, Kankel MW, Artavanis-Tsakonas S (2012) The notch signalling system: recent insights into the complexity of a conserved pathway. Nat Rev Genet 13:654–666
Zhang Z, Wang H (2020) lncRNA SNHG1 suppresses gastric cancer cell proliferation and promotes apoptosis via Notch1 pathway. J buon 25:302–307
Hu R, Lu Z (2020) Long non–coding RNA HCP5 promotes prostate cancer cell proliferation by acting as the sponge of miR–4656 to modulate CEMIP expression. Oncol Rep 43:328–336
Yin D, Lu X (2021) Silencing of long non-coding RNA HCP5 inhibits proliferation, invasion, migration, and promotes apoptosis via regulation of miR-299-3p/SMAD5 axis in gastric cancer cells. Bioengineered 12:225–239
Yang J, Antin P, Berx G, Blanpain C, Brabletz T, Bronner M et al (2020) Guidelines and definitions for research on epithelial–mesenchymal transition. Nat Rev Mol Cell Biol 21:341–352
Wu J, Liu T, Sun L, Zhang S, Dong G (2020) Long non-coding RNA SNHG4 promotes renal cell carcinoma tumorigenesis and invasion by acting as ceRNA to sponge mir-204-5p and upregulate RUNX2. Cancer Cell Int 20:1–14
Wang S, Zhu W, Qiu J, Chen F (2021) lncRNA SNHG4 promotes cell proliferation, migration, invasion and the epithelial-mesenchymal transition process via sponging mir-204-5p in gastric cancer. Mol Med Rep 23:1
Zhu M-B, Chen L-P, Hu M, Shi Z, Liu Y-N (2019) Effects of lncRNA BANCR on endometriosis through ERK/MAPK pathway. Eur Rev Med Pharmacol Sci. ;23
Shi Y, Sun H (2020) Down-regulation of lncRNA LINC00152 suppresses gastric cancer cell migration and invasion through inhibition of the ERK/MAPK signaling pathway. OncoTargets Therapy. :2115–2124
Zheng W, Li J, Zhou X, Cui L, Wang Y (2020) The lncRNA XIST promotes proliferation, migration and invasion of gastric cancer cells by targeting miR-337. Arab J Gastroenterol 21:199–206
Allavena P, Sica A, Garlanda C, Mantovani A (2008) The Yin-Yang of tumor‐associated macrophages in neoplastic progression and immune surveillance. Immunol Rev 222:155–161
Xie C, Guo Y, Lou S (2020) LncRNA ANCR promotes invasion and migration of gastric cancer by regulating FoxO1 expression to inhibit macrophage M1 polarization. Dig Dis Sci 65:2863–2872
Ayob AZ, Ramasamy TS (2018) Cancer stem cells as key drivers of tumour progression. J Biomed Sci 25:1–18
Zhang X, Hua R, Wang X, Huang M, Gan L, Wu Z et al (2016) Identification of stem-like cells and clinical significance of candidate stem cell markers in gastric cancer. Oncotarget 7:9815
Yao J, Yang Z, Yang J, Wang Z-G, Zhang Z-Y (2021) Long non-coding RNA FEZF1-AS1 promotes the proliferation and metastasis of hepatocellular carcinoma via targeting miR-107/Wnt/β-catenin axis. Aging 13:13726
Song B, Yan J, Liu C, Zhou H, Zheng Y (2015) Tumor suppressor role of mir-363-3p in gastric cancer. Med Sci Monitor: Int Med J Experimental Clin Res 21:4074
Hui Y, Yang Y, Li D, Wang J, Di M, Zhang S et al (2020) LncRNA FEZF1-AS1 modulates cancer stem cell properties of human gastric cancer through miR-363-3p/HMGA2. Cell Transplant 29:0963689720925059
Wang F, Tang C, Xu D, Tang Y, Jiang Y, Gao X et al (2020) LncRNA ADAMTS9-AS2 suppresses the proliferation of gastric cancer cells and the tumorigenicity of cancer stem cells through regulating SPOP. J Cell Mol Med 24:4830–4838
Liang G, He Z (2022) High mobility group proteins in sepsis. Front Immunol 13:911152
Xia Y, Lv J, Jiang T, Li B, Li Y, He Z et al (2021) CircFAM73A promotes the cancer stem cell-like properties of gastric cancer through the miR-490-3p/HMGA2 positive feedback loop and HNRNPK-mediated β-catenin stabilization. J Experimental Clin Cancer Res 40:1–24
Du T, Zhang B, Zhang S, Jiang X, Zheng P, Li J et al (2016) Decreased expression of long non-coding RNA WT1-AS promotes cell proliferation and invasion in gastric cancer. Biochimica et Biophysica Acta (BBA)-Molecular basis of Disease. 1862:12–19
Zhang X, Jin M, Liu S, Zang M, Hu L, Du T et al (2023) The roles and molecular mechanisms of long non-coding RNA WT1-AS in the maintenance and development of gastric cancer stem cells. Heliyon. ;9
Ma S, Xie N, Li W, Yuan B, Shi Y, Wang Y (2014) Immunobiology of mesenchymal stem cells. Cell Death Differ 21:216–225
Serra D, Mera P, Malandrino MI, Mir JF, Herrero L (2013) Mitochondrial fatty acid oxidation in obesity. Antioxid Redox Signal 19:269–284
Zhao L, Hantash BM (2011) TGF-β1 regulates differentiation of bone marrow mesenchymal stem cells. Vitamins Horm 87:127–141
He W, Liang B, Wang C, Li S, Zhao Y, Huang Q et al (2019) MSC-regulated lncRNA MACC1-AS1 promotes stemness and chemoresistance through fatty acid oxidation in gastric cancer. Oncogene 38:4637–4654
Acknowledgements
This study is supported via funding from Prince Sattam bin Abdulaziz University project number (PSAU/2023/R/1445).
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
R.S, S.A and A.H: Conceptualization, Writing - Original Draft. S.A and P.A: Conceptualization, Writing - Original Draft, Resources. A.H: Supervision, Project administration. T.A, M.J and A.A: Conceptualization, Writing - Review and Editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics statement
Not applicable.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Saleh, R.O., Al-Hawary, S.I.S., Hammoud, A. et al. The long non-coding RNAs (lncRNA) in the pathogenesis of gastric cancer cells: molecular mechanisms and involvement miRNAs. Mol Biol Rep 51, 615 (2024). https://doi.org/10.1007/s11033-024-09546-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11033-024-09546-x