Abstract
Background and Aim
Microscopic colitis, comprising collagenous colitis and lymphocytic colitis, is a common cause of chronic diarrhea. Previously, we showed enhanced chemokine productions in microscopic colitis patients, indicating dysregulated immune cell chemotaxis in the immunopathogenesis. We also showed decreased mRNA of IL-37, mainly regarded as an anti-inflammatory cytokine, in the colonic mucosa of these patients, potentially an important factor for the chronicity of the colitis. Our aim in this study was to understand the possible role of IL-37 in chemokine production using a cell line model.
Methods
A colon epithelial cell line, T84, was stimulated with the TLR5 ligand flagellin. IL-37 protein production was reduced 20% using the CRISPR/Cas9 system, and the changes in chemokine mRNA and protein expressions were compared to cells transfected with empty plasmid.
Results
The 20% reduction in IL-37 protein levels spontaneously increased CCL5, CXCL8, CXCL10, and CXCL11 mRNA and protein expressions. CCL2 mRNA and protein levels were enhanced upon TLR5 stimulation. CCL3, CCL20, and CX3CL1 mRNA expressions were increased either spontaneously or following TLR5 stimulation, whereas CCL4 and CCL22 mRNA expressions were significantly decreased.
Conclusions
Even a minor decrease in the ability of colon epithelial cells to produce IL-37 results in altered chemokine expression, mainly an increase in the production of several chemokines. Our results indicate that a decreased IL-37 expression by colon epithelial cells may be an important factor for increasing the recruitment of immune cells and subsequently developing microscopic colitis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Microscopic colitis (MC), comprising collagenous colitis (CC) and lymphocytic colitis (LC), is characterized clinically by chronic watery diarrhea. The diagnosis relies on typical histopathological features that are observed upon microscopic examination: lymphocytic infiltration of the epithelium and the lamina propria, as well as a damaged, flattened, and detached epithelial layer. CC patients have a characteristic thickened subepithelial collagen layer [1, 2]. Both CC and LC show a mixed inflammatory cell infiltration in the lamina propria, including T and B lymphocytes, plasma cells, eosinophils, neutrophils, mast cells, and macrophages [3–5]. Previously, we demonstrated upregulated gene (CCL2, 3, 4, 5, 22; CXCL8, 9, 10, 11; and CX3CL1) and protein (CCL2, 3, 4; CXCL8, 10; and CX3CL1) expressions in MC patients, which may be involved in recruiting these mixed cell populations as a part of the immunopathogenesis [6]. However, it is still not known what may trigger these aberrant chemokine expressions in MC patients.
In another study from our group, both CC and LC patients showed significantly decreased IL-37 gene expression in colonic biopsies, suggesting a potential factor involved in the chronicity of the colonic inflammation in these patients [7]. Expression of IL-37 can be induced by Toll-like receptor (TLR) agonists and pro-inflammatory cytokines such as IL-1β, tumor necrosis factor (TNF)-α, and interferon (IFN)-γ in peripheral blood mononuclear cells (PBMCs) and dendritic cells (DCs) [8, 9]. IL-37 is synthesized as a precursor and requires cleavage by caspase-1 to generate the mature form [10]. Mature IL-37 is either secreted extracellularly to inhibit the actions of pro-inflammatory cytokines or their receptors [11] or translocated to the nucleus, where it interacts with Smad3 and suppresses transcription of pro-inflammatory cytokines [12, 13]. In different cell types, IL-37 can effectively reduce production of TNF-α, macrophage inflammatory protein-2 (MIP-2, CXCL2), IL-1α, IL-6, macrophage colony-stimulating factor (M-CSF), B cell-attracting chemokine-1 (BCA-1, CXCL13), granulocyte–macrophage colony-stimulating factor (GM-CSF), IL-1β, monocyte chemoattractant protein-5 (MCP-5/CCL12), and CXCL8 upon IL-1/TLR stimulation [8, 12].
Although data on IL-37 continue to emerge in various disease states, knowledge of its functional role, particularly in colitis, is still in the early stages. Increased IL-37 protein levels in intestinal epithelial cells from patients with ulcerative colitis and Crohn’s disease have been detected using immunohistochemistry [14–17], whereas decreased IL-37 gene expression has been observed in MC patients in whole colonic biopsies [7]. Since the anti-inflammatory cytokine IL-37 has been suggested to be produced by not only several myeloid cells, but also epithelial cells in the colon [10, 14, 16], and was shown to be enhanced in the colonic epithelial cell line T84 upon stimulation with TNF-α [17], we focused on the possible impact of decreased mRNA levels of IL-37 on chemokine expression in the T84 cell line. To the best of our knowledge, there is no previous report on the importance of IL-37 in regulating chemokine expression in colon epithelial cells. In order to reduce IL-37 protein levels, we used the type II clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas) protein 9 (CRISPR/Cas9) system to achieve targeted mutagenesis in mammalian cells [18–20].
This study demonstrates the regulatory role of IL-37 on gene and protein expressions of chemokines, some of which are dysregulated in MC pathogenesis. Even a minor decrease in the ability of colon epithelial cells to produce IL-37 results in altered chemokine expression, mainly an increase in the production of several chemokines. Our results indicate that decreased IL-37 expression by colon epithelial cells may be an important factor in increasing the recruitment of immune cells and subsequently developing microscopic colitis.
Materials and Methods
Cell Culture
The human epithelial colorectal carcinoma cell line T84 (CCL248, American Type Culture Collection [ATCC], Manassas, VA, USA) [21] was cultured and subcultured according to ATCC instructions (http://www.lgcstandards-atcc.org/). Briefly, the cells were cultured at 37 °C under 5% CO2 in a 1:1 mixture of Ham’s F12 medium and Dulbecco’s modified Eagle’s medium (DMEM) with 2.5 mM l-glutamine (Gibco, Life Technologies, Grand Island, NY, USA) supplemented with 5% heat-inactivated fetal bovine serum (FBS, Gibco, Life Technologies) and 1% gentamicin (Gibco, Life Technologies). The cells reaching 70–90% confluence were detached using 0.5% trypsin–EDTA in PBS (Gibco, Life Technologies) and subcultured. The cells used in this study were from passages 10 to 21.
Flow Cytometric Analysis of Toll-Like Receptors (TLRs)
We performed flow cytometry to detect TLR2, 4, or 5 expression on the T84 cell surface. The cells were stained with mouse antihuman TLR2 conjugated with Alexa Fluor 488 (CD282, clone 11G7, BD Biosciences, Pharmingen, San Diego, USA), mouse antihuman TLR4 (CD284, clone 901, BioLegend, San Diego, CA, USA) conjugated with brilliant violet (BV) 786, and mouse antihuman TLR5 conjugated with Alexa Fluor 647 (CD285, clone 624915, BioLegend). Cells were also stained with isotype controls, mouse IgG1κ-Alexa Fluor 488 (BD Biosciences), IgG1κ-BV786 (BioLegend), or IgG1κ-Alexa Fluor 647 (BioLegend). The monocyte cell line THP-1 (TIB-202, ATCC) was used as a positive control for TLR2, 4, and 5 expressions on the cell surface [22] (a gift from Professor T. Bengtsson, School of Medical Sciences, Faculty of Medicine and Health, Örebro University). Data acquisition of at least 50,000 events was performed using a Gallios flow cytometer (Beckman Coulter Inc, Fullerton, CA, USA), and data analysis was performed using version 1.1 of the Kaluza software package (Beckman Coulter).
Total RNA Isolation, Reverse Transcription (RT), and Quantitative Real-Time Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted from T84 cells using a Gene MATRIX RNA/miRNA purification kit (EURx, Berlin, Germany) according to the manufacturer’s instructions and quantified using a NanoDrop 200 spectrophotometer (Thermo Scientific, Wilmington, DE, USA). All products used in the reverse transcription and qRT-PCR were ordered from Applied Biosystems (Life Technologies), and procedures were performed according to the manufacturer’s instructions. cDNA transcription from 500 ng total RNA was performed using a High-Capacity cDNA Reverse Transcription Kit. For gene expression analysis using qRT-PCR, a TaqMan Fast Universal Master Mix was used with the following TaqMan primer–probe sets [6, 7, 23]: IL-37 (Hs00367201_m1), CCL2/MCP-1 (Hs00234140_m1), CCL3/MIP-1α (Hs00234142_m1), CCL4/MIP-1β (Hs99999148_m1), CCL5/RANTES (Hs00174575_m1), CCL20/MIP3A (Hs00355476_m1), CCL22/MDC (Hs01574247_m1), CXCL8/IL-8 (Hs00174103_m1), CXCL9/MIG (Hs00171065_m1), CXCL10/IP-10 (Hs01124251_g1), CXCL11/I-TAC (Hs04187682_g1), and CX3CL1/fractalkine (Hs00171086_m1). Normalization of qRT-PCR results was performed using GAPDH (Hs99999905_m1), which showed the most stable results compared to 18S or GUSB in these cells. The samples were run in a Gene Bio-Rad CFX96 Touch Real-Time PCR Detection System (Bio-Rad Laboratories Inc, Hercules, CA, USA). The comparative threshold cycle method was used to compare samples with controls [24].
IL-37 and CXCL8 Detection Using Enzyme-Linked Immunosorbent Assays (ELISA)
Secreted CXCL8 or IL-37 protein levels in cell media were measured using CXCL8 or IL-37 DuoSet ELISA Development Systems, respectively (R&D Systems, Minneapolis, MN, USA) at 23 °C, and protein levels were calculated using a recombinant CXCL8 or IL-37 protein standard curve according to the manufacturer’s instructions.
To measure the intracellular production of IL-37, protein extraction from cells was performed in radio-immunoprecipitation assay (RIPA) buffer (Sigma-Aldrich, St Louis, MO, USA) containing a 1:100 protease inhibitor cocktail (catalogue no: P8340, Sigma-Aldrich) [6]. The detection of intracellular IL-37 from cell lysate was performed using a human IL-37 ELISA Kit (Adipogen, Life Sciences, Liestal, Switzerland) due to higher sensitivity (10 pg/ml). Protein levels were calculated using a recombinant IL-37 protein standard curve according to the manufacturer’s instructions.
Optimization of Flagellin Stimulation
To detect the optimal concentration of TLR5 stimulation with its ligand flagellin, recombinant flagellin from Salmonella typhimurium (Novus Biologicals, Cambridge, UK) was used [25]. T84 cells were cultured at 50,000 cells/cm2 until they reached 70–90% confluence (approximately the fourth day of culture) [26] and then stimulated for 24 h with a series of flagellin concentrations: 10, 50, 100, or 500 ng/ml in culture media without FBS or antibiotics at 37 °C under 5% CO2. At the end of the incubation, cells and culture media were collected for further gene and protein expression analyses of IL-37 and control of TLR5 response via CXCL8 [27]. According to the results from the 24 h flagellin stimulation, the optimal stimulation time was further analyzed for 6, 12, or 48 h using the minimum (10 ng/ml) or the optimal (100 ng/ml) flagellin stimulation and the optimum TLR5 response was analyzed as described above.
Reduction in IL-37 Expression Using the CRISPR/Cas9 System
Single guide RNA (sgRNA), specific to the target site of IL-37a-e, was designed using the E-CRISP software (http://www.e-crisp.org/E-CRISP/) [28]. The target sequence (sgRNA) was cloned into the CRISPR/Cas9 plasmid backbone using a previously described protocol [29]. During the optimizations of the CRISPR/Cas9 system, we constructed two self-ligated empty plasmid controls using a Px459 plasmid (pSpCas9(BB)-2A-Puro (PX459) version 2.0, a gift from Feng Zhang, Addgene 62988) to allow self-ligation, as well as six IL-37sgRNA containing plasmids. Of these six plasmids, two showed similar results based on Western blot in reduction in IL-37 protein levels. For consistency, we chose one clone each for our subsequent analyses. Briefly, forward (100 µM, 5′–3′ CACCGTCCTGAGTTCTCCCCCACAA) and reverse (100 µM, 5′–3′ AAACTTGTGGGGGAGAACTCAGGAC) primers were annealed using T4 polynucleotide kinase (NEB, New England Biolabs Inc, Ipswich, MA, USA). The Px459 plasmid was digested overnight using the site specific BbsI enzyme (NEB). The ligation of annealed sgRNAs and Px459 plasmid was performed using T4 DNA ligase (Thermo Fischer Scientific, Wilmington, DE, USA). Chemically competent TOP10 E. coli (Invitrogen, Thermo Fischer Scientific) was used to transform the ligated plasmids. The plasmids were isolated using a QIAprep Spin Miniprep Kit (Qiagen, Hilden, Germany) and sent for sequencing to Eurofins Genomics Sequencing (Ebersberg, Germany).
The cells were transfected with 2 μg of IL-37sgRNA or an empty plasmid (TFneg) using an Amaxa Cell Line Nucleofector Kit T for T84 cells (Lonza, Cologne, Germany) in a Nucleofector II Device (Lonza). After 48 h of transfection, IL-37sgRNA and TFneg cells were treated with 4 μg/ml puromycin (Sigma-Aldrich) to select transfected cells. Optimized flagellin stimulation was then repeated for IL-37sgRNA treated and TFneg cells (passages 6 and 7), after which cells and culture media were collected for further analysis.
Western Blot
The protein concentrations of the cell lysate were determined using a DC Protein Assay Kit (Bio-Rad). To detect the expression of IL-37 in IL-37sgRNA and TFneg cells, 50 μg total protein from cell lysates was resolved in 12% Bis/Tris gels (Novex, Life Technologies) in NuPage running buffer (Novex, Life Technologies) and transferred to nitrocellulose membranes in blotting buffer (Bio-Rad). After blocking in 5% bovine serum albumin (BSA, Carl Roth GmbH, Karlsruhe, Germany), nitrocellulose membranes were probed overnight at 4 °C using 3 µg/ml rabbit polyclonal anti-IL-37b (Novus Biologicals, Cambridge, UK). Rabbit polyclonal anti-GAPDH (Santa Cruz Biotechnology, Dallas, Texas) at a 1:15,000 dilution was used as a loading control. Blots were then incubated with a horseradish peroxidase-conjugated secondary anti-rabbit IgG (Santa Cruz Biotechnology, Dallas, TX, USA) and visualized using a Luminata Forte Western HRP substrate (Merck Millipore, Billerica, MA, USA). The membranes were exposed to chemiluminescence using an Odyssey Fc (LI-COR Biosciences, Lincoln, NE, USA). Densitometric analysis was performed using version 3.1 of the Image Studio software package (LI-COR). The calculations were performed based on density of the IL-37 and GAPDH bands, respectively, as detected by the software, and the results were normalized based on IL-37/GAPDH.
Luminex
The cells (wild type, IL-37sgRNA, and TFneg cells) were incubated for 12 h either with or without 100 ng/ml flagellin in cell culture media without FBS or gentamicin. The cell culture media were then collected, aliquoted, and stored at −80 °C until further analysis. Protein levels of CCL2, 3, 4, 5, 20, 22; CXCL9, 10, 11; and CX3CL1 from 25 μl cell culture media were analyzed in duplicates by xMAP technology developed using the Luminex (Austin, TX, USA) [6]. The samples were incubated with chemokine antibody-immobilized beads overnight, then with detection antibodies for 1 h, and finally for 30 min after the direct addition of streptavidin–phycoerythrin, all at room temperature. After subsequent washes, 150 μl wash buffer was added to samples for data acquisition. The concentrations were determined in a Luminex 200 (Millipore, MA, USA) using a Milliplex Map Kit (Millipore) according to the manufacturer’s instructions. The levels of different chemokines from samples and controls were expressed as pg/ml, according to a standard curve with known amounts of each analyte.
Statistical Analysis
All data are shown as mean ± standard error of the mean (SEM). For optimization of flagellin stimulation, the results from flagellin-stimulated cells were compared with unstimulated T84 cells. The results from the cells transfected with CRISPR/Cas9 plasmid containing IL-37sgRNA (IL-37sgRNA cells) were compared with the results from the cells transfected with an empty CRISPR/Cas9 plasmid (TFneg cells), and the unstimulated cells were compared to stimulated cells within the same cell type (wild type [WT], TFneg, or IL-37sgRNA). All comparisons were performed using analysis of variance (ANOVA) with Tukey’s multiple comparisons test; statistical significance was set at p ≤ 0.05 (GraphPad Prism 5, San Diego, CA, USA). Statistically significant differences are marked with *, **, or ***, depicting p < 0.05, <0.01, or <0.001.
Results
Optimization of TLR5 Response in Intestinal Epithelial Cells Stimulated with Flagellin
We detected TLR2, 4, and 5 expression on the THP-1 cell surface, whereas the majority of T84 cells expressed TLR5 on their surface (data not shown), in line with previous studies [30]. Neither TLR2 nor TLR4 expression was detected on T84 cells (data not shown) [31]. The experiments were therefore performed using recombinant flagellin from Salmonella typhimurium, a TLR5 ligand. The optimum flagellin stimulation was chosen according to protein levels of IL-37 from cell lysates and CXCL8 from cell media; the latter was used as an indicator for TLR5 signaling due to a strong induction of CXCL8 upon flagellin stimulation [27]. Following 24 h stimulation, our results showed that 100 ng/ml flagellin was the minimum amount that significantly induced IL-37 mRNA expression (data not shown) and increased protein expression, although it did not reach significance compared to unstimulated cells (Fig. 1a). Thereafter, according to results from the 24 h stimulation, the cells were stimulated with 10 or 100 ng/ml for 6, 12, or 48 h (Fig. 1a). Significant increases in intracellular IL-37 protein levels were observed in cells stimulated with 10 or 100 ng/ml flagellin for 12 h (Fig. 1a). In addition, 100 ng/ml flagellin stimulation for 12 h caused a significant increase in IL-37 mRNA expression compared to unstimulated cells (data not shown).
IL-37 and CXCL8 protein expressions in response to 10 or 100 ng/ml flagellin stimulation of T84 cells for different time periods. Protein levels of intracellular IL-37 (a), secreted IL-37 (b), and secreted CXCL8 (c) in cell culture media were compared to unstimulated controls (n = 4). Statistically significant differences are marked as *, **, or *** and depict p < 0.05, < 0.01, or < 0.001, respectively. All data are shown as mean ± SEM
It has previously been shown that upon TLR signaling, IL-37 can be secreted extracellularly to inhibit expression of cytokines, such as IFN-γ [11, 32]. We also addressed whether secreted IL-37 could be involved in regulation of chemokine expressions. IL-37 protein levels from cell media were significantly increased following stimulation with 100 ng/ml flagellin for 6 h, albeit not at 12, 24, or 48 h, compared to unstimulated cells (Fig. 1b). CXCL8 protein levels in cell media were significantly enhanced when stimulated with two different flagellin concentrations (100 and 500 ng/ml) after 24 h (data not shown). In addition, only 100 ng/ml flagellin stimulation led to increased CXCL8 protein levels in cell media after all four time points (Fig. 1c). Therefore, according to the mRNA and protein expression results of IL-37 from cell lysate and CXCL8 from cell media, 100 ng/ml flagellin stimulation for 12 h was chosen as an optimum setting for further experiments.
IL-37 Protein Levels Were Reduced by 20% Using the CRISPR/Cas9 System Compared to Cells Transfected with Empty CRISPR/Cas9 Plasmid
The CRISPR/Cas9 mediated reduction in IL-37 protein levels was assessed by Western blot (Fig. 2). Densitometric measurements from the Western blot where IL-37 protein levels were normalized to GAPDH protein levels demonstrated a 20% reduced protein expression in comparison with TFneg cells.
Reduction in IL-37 protein expression detected by Western blot. CRISPR/Cas9 mediated reduction in IL-37 protein expression. T84 cells were transfected with IL-37sgRNA and intracellular IL-37 protein expressions were compared to cells transfected with an empty plasmid (TFneg). GAPDH served as a loading control. 50 μg of total protein from whole cell lysate was loaded. WT wild-type T84 cells
Reduction in IL-37 Protein Levels Significantly Increased mRNA and Protein Expressions of CCL2, CCL5, CXCL8, CXCL10, and CXCL11
CCL2 is an important chemoattractant for monocytes and plays a role in T helper type 2 responses [33–35]. IL-37sgRNA cells showed significant increases in CCL2 mRNA (Fig. 3a) and protein (Fig. 3b) expressions upon flagellin stimulation compared to both unstimulated IL-37sgRNA cells and compared to flagellin-stimulated TFneg cells, whereas unstimulated IL-37sgRNA cells showed significantly decreased CCL2 protein levels compared to unstimulated TFneg cells (Fig. 3b).
mRNA (a) and protein (b) levels of CCL2, CCL5, CXCL8, CXCL10, and CXCL11 (n = 5). The filled bars represent cells stimulated with 100 ng/ml flagellin for 12 h, whereas empty bars denote unstimulated cells. First, unstimulated cells with reduced IL-37 levels (IL-37sgRNA) were compared to unstimulated cells transfected with an empty plasmid (TFneg), whereas the IL-37sgRNA cells stimulated with flagellin are compared to TFneg cells stimulated with flagellin. Second, unstimulated cells are compared to flagellin-stimulated cells, within the same cell type (WT, TFneg or IL-37sgRNA). Statistically significant differences are marked with *, **, or *** depicting p < 0.05, < 0.01, or < 0.001. All data are shown as mean ± SEM. WT wild-type T84 cells, N.D not detected
CCL5 is a pleiotropic chemokine attracting Treg cells, eosinophils, and macrophages [36, 37], whereas CXCL8 selectively recruits neutrophils [36]. IL-37sgRNA cells showed significantly increased CCL5 mRNA expression compared to TFneg cells, as well as spontaneously increased protein levels (Fig. 3a, b). CXCL8 mRNA and protein levels were significantly upregulated upon flagellin stimulation in IL-37sgRNA cells compared to unstimulated IL-37sgRNA cells (Fig. 3a, b). CXCL8 mRNA levels were also significantly increased upon flagellin stimulation in WT cells, and protein levels were only detected in flagellin-stimulated WT cells (Fig. 3a, b). Furthermore, flagellin-stimulated IL-37sgRNA cells showed significantly increased CXCL8 mRNA and protein levels compared to flagellin-stimulated TFneg cells (Fig. 3a, b). Similarly, chemokine recruiting T helper 1 (Th1) and CD8+ T cells CXCL11 [38] had significant increases in both mRNA and protein levels in IL-37sgRNA cells compared to TFneg cells, regardless of flagellin stimulation (Fig. 3a, b). CXCL10 mRNA levels significantly increased upon flagellin stimulation in IL-37sgRNA cells compared to TFneg cells. Moreover, flagellin stimulation led to a significant increase in CXCL10 mRNA expression in WT and IL-37sgRNA cells compared to their unstimulated counterparts (Fig. 3a). Additionally, the reduction in IL-37 protein levels led to spontaneous increase in mRNA and protein levels of CCL5, CXCL8, CXCL10, and CXCL11 (Fig. 3a, b).
Reduced IL-37 Protein Levels Significantly Increased CCL3, CCL20, and CX3CL1 mRNA Expressions, Whereas CCL4 and CCL22 mRNA Expressions Were Significantly Decreased
CCL3 attracts macrophages [36, 39], whereas CX3CL1 recruits macrophages, Th1 and CD8+ T cells [36]. In IL-37sgRNA cells, the reduced IL-37 protein levels caused significantly increased CCL3 and CX3CL1 mRNA expressions compared to TFneg cells, which was not changed upon flagellin stimulation (Fig. 4a). However, protein levels of CCL3 were lower than the detection limit in all cell types (3.2 pg/ml, Fig. 4b). Compared to TFneg cells, CX3CL1 protein levels were significantly decreased in IL-37sgRNA cells, regardless of flagellin stimulation (Fig. 4b).
mRNA (a) and protein (b) levels of CCL3, CX3CL1, and CCL20 (n = 5). The filled bars represent cells stimulated with 100 ng/ml flagellin for 12 h, whereas empty bars denote unstimulated cells. First, unstimulated cells with reduced IL-37 levels (IL-37sgRNA) were compared to unstimulated cells transfected with an empty plasmid (TFneg), whereas the IL-37sgRNA cells stimulated with flagellin are compared to TFneg cells stimulated with flagellin. Second, unstimulated cells are compared to flagellin-stimulated cells, within the same cell type (WT, TFneg or IL-37sgRNA). Statistically significant differences are marked with *, **, or *** depicting p < 0.05, < 0.01, or < 0.001. All data are shown as mean ± SEM. WT wild-type T84 cells, N.D not detected
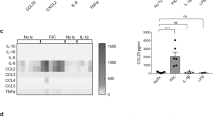
CCL20 selectively recruits Th17 cells [36] and has previously been shown to be upregulated by flagellin stimulation [27, 40]. In the present study, flagellin stimulation significantly upregulated CCL20 mRNA expression in all cell types compared to unstimulated cells (Fig. 4a). Similarly, flagellin stimulation in WT cells led to significantly increased CCL20 protein levels compared to unstimulated WT cells (Fig. 4b). CCL20 mRNA levels were significantly enhanced in flagellin-stimulated IL-37sgRNA cells compared to flagellin-stimulated TFneg cells (Fig. 4a), whereas CCL20 protein levels in cell media were not significantly changed (Fig. 4b).
CCL4 and CCL22 attract macrophages [36, 39], and CCL4 can also recruit eosinophils [36, 37]. Upon flagellin stimulation, all cell types showed significantly increased CCL22 mRNA levels compared to unstimulated cells, whereas IL-37sgRNA cells showed significantly reduced CCL4 and CCL22 mRNA expressions compared to TFneg cells (Fig. 5a). CCL4 protein levels were lower than the detection limit (3.2 pg/ml), and no significant change was detected in CCL22 protein levels in cell media (Fig. 5b).
mRNA (a) and protein (b) levels of CCL4, CCL22, and CXCL9 (n = 5). The filled bars represent cells stimulated with 100 ng/ml flagellin for 12 h, whereas empty bars denote unstimulated cells. First, unstimulated cells with reduced IL-37 levels (IL-37sgRNA) were compared to unstimulated cells transfected with an empty plasmid (TFneg), whereas the IL-37sgRNA cells stimulated with flagellin are compared to TFneg cells stimulated with flagellin. Second, unstimulated cells are compared to flagellin-stimulated cells, within the same cell type (WT, TFneg or IL-37sgRNA). Statistically significant differences are marked with *, **, or *** depicting p < 0.05, < 0.01, or < 0.001. All data are shown as mean ± SEM. WT wild-type T84 cells, N.D not detected
CXCL9 recruits Th1 and CD8+ T cells [38]. In WT cells, flagellin stimulation led to significantly increased CXCL9 mRNA expression compared to unstimulated WT cells, and CXCL9 protein levels were detected only in flagellin-stimulated WT cells (Fig. 5a, b). We did not detect any CXCL9 protein or any significant changes in CXCL9 mRNA expression in IL-37sgRNA or TFneg cells regardless of flagellin stimulation (Fig. 5a, b).
Discussion
In this study, we used an intestinal epithelial cell line model to understand the possible regulatory role of IL-37 on chemokines involved in recruitment of immune cells in the immunopathology of MC. Even a minor reduction in IL-37 protein levels resulted in noticeable upregulations in mRNA and protein expressions of CCL2, CCL5, CXCL8, and CXCL11. In addition, IL-37 may be involved in regulation of CCL3, CXCL10, CX3CL1, and CCL20 mRNA expressions, whereas CCL4 and CCL22 mRNA expressions were further decreased in cells with reduced IL-37 protein levels. Overall, our results show that IL-37 might be an important regulatory factor involved in chemokine expressions by colon epithelial cells and could play a role in the development of MC.
The intestinal epithelial cell line T84 expressed TLR5 on the cell surface, in line with previous findings [31]. Colonic epithelial cells express TLR5 predominantly on the basolateral side, which is separated from the colon microbiota [41]. Due to epithelial damage in inflammatory bowel disease (IBD) patients, the bacteria can reach the basolateral side of epithelial cells and may lead to aberrant immune responses [42], which is supported through upregulation of TLR5 expression in IBD patients [43]. Similarly, MC patients also show intestinal epithelial cell damage [1] and enhanced production of CCL2 [6]. In IBD patients, increased CCL2 mRNA and protein expressions were detected in areas with epithelial cell damage and were correlated with disease activity [44]. Enhanced CCL2 mRNA and protein levels in cells with reduced IL-37 levels indicate a possible correlation between TLR5 stimulation due to enhanced contact with microbiota and CCL2 expressions in disease immunopathogenesis. In addition, the present study shows that TLR5 signaling led to increased mRNA and protein levels, corroborating our previous studies [6, 23] as well as findings in Crohn’s disease (CD) and ulcerative colitis (UC) patients [44–49].
IL-37 gene and protein expressions were induced by TLR5 signaling. As we observed no significant changes in IL-37 secretion in the cell media of T84 cells after 12 h of flagellin stimulation, the main focus of this study was the function of intracellular IL-37, which suppresses transcription of cytokines and chemokines in the nucleus [12, 13]. Spontaneous increases in chemokine mRNA and protein expressions in cells with reduced IL-37 protein levels support an anti-inflammatory role of intracellular IL-37 on chemokine production [12, 13]. Upregulation of these chemokines might be due to the diminished IL-37 gene levels observed in MC patients [7], which in turn may contribute to their aberrant immune responses via recruiting mixed immune cell populations of neutrophils, eosinophils, monocytes, and effector T cells [36, 37, 50, 51]. The sum of a changed chemokine pattern due to reduced IL-37 expression warrants further functional studies aimed at dissecting MC disease pathology using a transwell migration assay to determine the effect of reduced IL-37 expression in T84 cells in attracting immune cells.
We suggest that the 20% reduction in IL-37 expression compared to TFneg cells, which might be considered a modest effect, may in fact better mirror the in vivo situation and the changes observed in CC and LC patients ex vivo due to a reduction in IL-37 gene expression rather than a complete loss of function in these patients [7]. Our previous study showing a similar chemokine expression profile [6] further supports our current model of a reduced IL-37 production which affects chemokine production. MC is a chronic inflammatory disease, with the highest incidence in people > 60 years. It has been shown that elderly people can have altered cytokine productions which affects the inflammatory response [reviewed in [52, 53]]. This cell line model with reduced IL-37 protein levels may suggest a possible risk of MC development in elderly people, if the ability to produce IL-37 decreases in certain individuals [53].
Decreased IL-37 protein levels in T84 cells were achieved by targeted mutagenesis using the CRISPR/Cas9 system [19], in which Cas9 protein triggered immune responses or feedback mechanisms in T84 cells. To exclude possible effects of transfection on chemokine expression, IL-37sgRNA cells were compared to cells transfected with plasmids without IL-37sgRNA constructs (TFneg cells). Although we achieved a minor reduction (20%) using IL-37sgRNA, the changes in chemokine expression due to reduced IL-37 protein levels were evident for several chemokines. These reductions may be clinically more relevant for understanding MC immunopathogenesis, since these patients demonstrated reduced IL-37 gene expressions but not a defect in the gene level [7]. These results may elucidate the effect of reduced IL-37 mRNA and protein levels observed also in other inflammatory diseases, such as Behҫet’s disease [54], ulcerative colitis [7], and hepatocellular carcinoma [55].
Conclusions
In this study, we have demonstrated the regulatory role of IL-37 on chemokine gene and protein expressions in the colon epithelial cell line. Our results show that even a minor decrease in the ability of colon epithelial cells to produce IL-37 results in a change in chemokine expression, mainly an increase in the production of several chemokines. A decreased IL-37 expression by colon epithelial cells may therefore be an important factor in increasing the recruitment of immune cells and subsequently developing microscopic colitis.
References
Bohr J, Wickbom A, Hegedus A, Nyhlin N, Hultgren Hörnquist E, Tysk C. Diagnosis and management of microscopic colitis: current perspectives. Clin Exp Gastroenterol. 2014;7:273–284.
Mahajan D, Goldblum JR, Xiao SY, Shen B, Liu X. Lymphocytic colitis and collagenous colitis: a review of clinicopathologic features and immunologic abnormalities. Adv Anat Pathol. 2012;19:28–38.
Nishida Y, Murase K, Isomoto H, et al. Different distribution of mast cells and macrophages in colonic mucosa of patients with collagenous colitis and inflammatory bowel disease. Hepatogastroenterology. 2002;49:678–682.
Levy AM, Yamazaki K, Van Keulen VP, et al. Increased eosinophil infiltration and degranulation in colonic tissue from patients with collagenous colitis. Am J Gastroenterol. 2001;96:1522–1528.
Göranzon C, Kumawat AK, Hultgren-Hörnqvist E, et al. Immunohistochemical characterization of lymphocytes in microscopic colitis. J Crohns Colitis.. 2013;7:e434–e442.
Günaltay S, Kumawat AK, Nyhlin N, et al. Enhanced levels of chemokines and their receptors in the colon of microscopic colitis patients indicate mixed immune cell recruitment. Mediators Inflamm. 2015;2015:132458.
Günaltay S, Nyhlin N, Kumawat AK, et al. Differential expression of interleukin-1/Toll-like receptor signaling regulators in microscopic and ulcerative colitis. World J Gastroenterol. 2014;20:12249–12259.
Xu WD, Zhao Y, Liu Y. Insights into IL-37, the role in autoimmune diseases. Autoimmun Rev. 2015;14:1170–1175.
Bufler P, Gamboni-Robertson F, Azam T, Kim SH, Dinarello CA. Interleukin-1 homologues IL-1F7b and IL-18 contain functional mRNA instability elements within the coding region responsive to lipopolysaccharide. Biochem J. 2004;381:503–510.
Kumar S, Hanning CR, Brigham-Burke MR, et al. Interleukin-1F7B (IL-1H4/IL-1F7) is processed by caspase-1 and mature IL-1F7B binds to the IL-18 receptor but does not induce IFN-gamma production. Cytokine. 2002;18:61–71.
Bulau AM, Nold MF, Li S, et al. Role of caspase-1 in nuclear translocation of IL-37, release of the cytokine, and IL-37 inhibition of innate immune responses. Proc Natl Acad Sci USA. 2014;111:2650–2655.
Nold MF, Nold-Petry CA, Zepp JA, Palmer BE, Bufler P, Dinarello CA. IL-37 is a fundamental inhibitor of innate immunity. Nat Immunol. 2010;11:1014–1022.
Sharma S, Kulk N, Nold MF, et al. The IL-1 family member 7b translocates to the nucleus and down-regulates proinflammatory cytokines. J Immunol. 2008;180:5477–5482.
Li Y, Wang Y, Liu Y, Zuo X, Lu X. The possible role of the novel cytokines IL-35 and IL-37 in inflammatory bowel disease. Mediat Inflamm. 2014;2014:136329.
Fonseca-Camarillo G, Furuzawa-Carballeda J, Yamamoto-Furusho JK. Interleukin 35 (IL-35) and IL-37: intestinal and peripheral expression by T and B regulatory cells in patients with inflammatory bowel disease. Cytokine. 2015;75:389–402.
Weidlich S, Bulau AM, Schwerd T, et al. Intestinal expression of the anti-inflammatory interleukin-1 homologue IL-37 in pediatric inflammatory bowel disease. J Pediatr Gastroenterol Nutr. 2014;59:e18–e26.
Imaeda H, Takahashi K, Fujimoto T, et al. Epithelial expression of interleukin-37b in inflammatory bowel disease. Clin Exp Immunol.. 2013;172:410–416.
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821.
Zhang F, Wen Y, Guo X. CRISPR/Cas9 for genome editing: progress, implications and challenges. Hum Mol Genet. 2014;23:R40–R46.
Shalem O, Sanjana NE, Zhang F. High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet. 2015;16:299–311.
Dharmsathaphorn K, McRoberts JA, Mandel KG, Tisdale LD, Masui H. A human colonic tumor cell line that maintains vectorial electrolyte transport. Am J Physiol. 1984;246:G204–G208.
Verma R, Jung JH, Kim JY. 1,25-Dihydroxyvitamin D3 up-regulates TLR10 while down-regulating TLR2, 4, and 5 in human monocyte THP-1. J Steroid Biochem Mol Biol. 2014;141:1–6.
Kumawat AK, Strid H, Tysk C, Bohr J, Hörnquist EH. Microscopic colitis patients demonstrate a mixed Th17/Tc17 and Th1/Tc1 mucosal cytokine profile. Mol Immunol. 2013;55:355–364.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408.
Gillaux C, Méhats C, Vaiman D, Cabrol D, Breuiller-Fouché M. Functional screening of TLRs in human amniotic epithelial cells. J Immunol.. 2011;187:2766–2774.
Watson RO, Galán JE. Campylobacter jejuni survives within epithelial cells by avoiding delivery to lysosomes. PLoS Pathog. 2008;4:e14.
Bannon C, Davies PJ, Collett A, Warhurst G. Potentiation of flagellin responses in gut epithelial cells by interferon-gamma is associated with STAT-independent regulation of MyD88 expression. Biochem J. 2009;423:119–128.
Heigwer F, Kerr G, Boutros M. E-CRISP: fast CRISPR target site identification. Nat Methods. 2014;11:122–123.
Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–784.
Harrison LM, Rallabhandi P, Michalski J, et al. Vibrio cholerae flagellins induce Toll-like receptor 5-mediated interleukin-8 production through mitogen-activated protein kinase and NF-kappaB activation. Infect Immun. 2008;76:5524–5534.
van Aubel RA, Keestra AM, Krooshoop DJ, van Eden W, van Putten JP. Ligand-induced differential cross-regulation of Toll-like receptors 2, 4 and 5 in intestinal epithelial cells. Mol Immunol. 2007;44:3702–3714.
Bufler P, Azam T, Gamboni-Robertson F, et al. A complex of the IL-1 homologue IL-1F7b and IL-18-binding protein reduces IL-18 activity. Proc Natl Acad Sci U S A.. 2002;99:13723–13728.
Serbina NV, Pamer EG. Monocyte emigration from bone marrow during bacterial infection requires signals mediated by chemokine receptor CCR2. Nat Immunol.. 2006;7:311–317.
Sozzani S, Zhou D, Locati M, et al. Receptors and transduction pathways for monocyte chemotactic protein-2 and monocyte chemotactic protein-3. Similarities and differences with MCP-1. J Immunol. 1994;152:3615–3622.
Gu L, Tseng S, Horner RM, Tam C, Loda M, Rollins BJ. Control of TH2 polarization by the chemokine monocyte chemoattractant protein-1. Nature. 2000;404:407–411.
Bachelerie F, Ben-Baruch A, Burkhardt AM, et al. International union of basic and clinical pharmacology. LXXXIX. Update on the extended family of chemokine receptors and introducing a new nomenclature for atypical chemokine receptors. Pharmacol Rev. 2014;66:1–79.
Hogan SP, Rothenberg ME, Forbes E, Smart VE, Matthaei KI, Foster PS. Chemokines in eosinophil-associated gastrointestinal disorders. Curr Allergy Asthma Rep. 2004;4:74–82.
Groom JR, Luster AD. CXCR3 ligands: redundant, collaborative and antagonistic functions. Immunol Cell Biol. 2011;89:207–215.
Mantovani A, Gray PA, Van Damme J, Sozzani S. Macrophage-derived chemokine (MDC). J Leukoc Biol. 2000;68:400–404.
Yoshino Y, Kitazawa T, Ikeda M, et al. Clostridium difficile flagellin stimulates toll-like receptor 5, and toxin B promotes flagellin-induced chemokine production via TLR5. Life Sci. 2013;92:211–217.
Abreu MT. Toll-like receptor signalling in the intestinal epithelium: how bacterial recognition shapes intestinal function. Nat Rev Immunol. 2010;10:131–144.
Lodes MJ, Cong Y, Elson CO, et al. Bacterial flagellin is a dominant antigen in Crohn disease. J Clin Invest. 2004;113:1296–1306.
Cario E, Podolsky DK. Differential alteration in intestinal epithelial cell expression of toll-like receptor 3 (TLR3) and TLR4 in inflammatory bowel disease. Infect Immun. 2000;68:7010–7017.
Banks C, Bateman A, Payne R, Johnson P, Sheron N. Chemokine expression in IBD. Mucosal chemokine expression is unselectively increased in both ulcerative colitis and Crohn’s disease. J Pathol. 2003;199:28–35.
Skovdahl HK, Granlund A, Østvik AE, et al. Expression of CCL20 and its corresponding receptor CCR6 is enhanced in active inflammatory bowel disease, and TLR3 mediates CCL20 expression in colonic epithelial cells. PLoS One. 2015;10:e0141710.
Jugde F, Alizadeh M, Boissier C, et al. Quantitation of chemokines (MDC, TARC) expression in mucosa from Crohn’s disease and ulcerative colitis. Eur Cytokine Netw. 2001;12:468–477.
Kvedaraite E, Lourda M, Ideström M, et al. Tissue-infiltrating neutrophils represent the main source of IL-23 in the colon of patients with IBD. Gut. 2016;65:1632–1641.
Egesten A, Eliasson M, Olin AI, et al. The proinflammatory CXC-chemokines GRO-alpha/CXCL1 and MIG/CXCL9 are concomitantly expressed in ulcerative colitis and decrease during treatment with topical corticosteroids. Int J Colorectal Dis. 2007;22:1421–1427.
Uguccioni M, Gionchetti P, Robbiani DF, et al. Increased expression of IP-10, IL-8, MCP-1, and MCP-3 in ulcerative colitis. Am J Pathol. 1999;155:331–336.
Pelletier M, Maggi L, Micheletti A, et al. Evidence for a cross-talk between human neutrophils and Th17 cells. Blood. 2010;115:335–343.
Zimmerman NP, Vongsa RA, Wendt MK, Dwinell MB. Chemokines and chemokine receptors in mucosal homeostasis at the intestinal epithelial barrier in inflammatory bowel disease. Inflamm Bowel Dis. 2008;14:1000–1011.
Agarwal S, Busse PJ. Innate and adaptive immunosenescence. Ann Allergy Asthma Immunol.. 2010;104:183–190. quiz 190–182, 210.
Mabbott NA, Kobayashi A, Sehgal A, Bradford BM, Pattison M, Donaldson DS. Aging and the mucosal immune system in the intestine. Biogerontology. 2015;16:133–145.
Ye Z, Wang C, Kijlstra A, Zhou X, Yang P. A possible role for interleukin 37 in the pathogenesis of Behcet’s disease. Curr Mol Med.. 2014;14:535–542.
Zhao JJ, Pan QZ, Pan K, et al. Interleukin-37 mediates the antitumor activity in hepatocellular carcinoma: role for CD57+ NK cells. Sci Rep. 2014;4:5177.
Acknowledgments
This study was supported by grants from the Swedish Society of Medicine (Bengt Ihre Foundation, Grants SLS-176271/2011, 98031/2010), the Örebro University Hospital Research Foundation (Nyckelfonden), and the Örebro County Council Research Committee. Sezin Günaltay’s salary is covered by a grant from Örebro University. We are very grateful to Maximilian W. Schenk at the Division of Signaling and Functional Genomics, German Cancer Research Center (DKFZ), Heidelberg, Germany, for help with the CRISPR/Cas9 system experimental setup and optimizations.
Author’s Contribution
SG and MG carried out the experiments, optimized the flagellin stimulations, performed the data analysis, and drafted the manuscript. EHH, OH, and SG created the study design, coordination, and data analysis. EHH, OH, and SG finalized the manuscript. All authors read and approved the final manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest regarding the publication of this article.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/), which permits any noncommercial use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Günaltay, S., Ghiboub, M., Hultgren, O. et al. Reduced IL-37 Production Increases Spontaneous Chemokine Expressions in Colon Epithelial Cells. Dig Dis Sci 62, 1204–1215 (2017). https://doi.org/10.1007/s10620-016-4422-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10620-016-4422-9