Abstract
Functional connectivity (i.e., the movement of individuals across a landscape) is essential for the maintenance of genetic variation and persistence of rare species. However, illuminating the processes influencing functional connectivity and ultimately translating this knowledge into management practice remains a fundamental challenge. Here, we combine various population structure analyses with pairwise, population-specific demographic modeling to investigate historical functional connectivity in Graham’s beardtongue (Penstemon grahamii), a rare plant narrowly distributed across a dryland region of the western US. While principal component and population structure analyses indicated an isolation-by-distance pattern of differentiation across the species’ range, spatial inferences of effective migration exposed an abrupt shift in population ancestry near the range center. To understand these seemingly conflicting patterns, we tested various models of historical gene flow and found evidence for recent admixture (~ 3400 generations ago) between populations near the range center. This historical perspective reconciles population structure patterns and suggests management efforts should focus on maintaining connectivity between these previously isolated lineages to promote the ongoing transfer of genetic variation. Beyond providing species-specific knowledge to inform management options, our study highlights how understanding demographic history may be critical to guide conservation efforts when interpreting population genetic patterns and inferring functional connectivity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Intensifying interactions between habitat destruction and climate change are a major threat to the persistence of many native species across the globe (Travis 2004; Mantyka-Pringle et al. 2012). As these processes fragment a species’ range, isolated populations may experience stronger genetic drift and inbreeding, leading to the loss of genetic diversity and increasing the chance of local extinction over time (Wright 1931; Allendorf et al. 2012). Species with genetically diverse and interconnected populations are likely more resilient to environmental change (Fisher 1930; Sgro et al. 2011) and may ultimately contribute to more stable communities and ecosystems (Hughes et al. 2008). Maintaining functional connectivity (i.e., the movement of individuals and genes across the landscape; Crooks & Sanjayan, 2006) between populations of rare species in the face of increasing habitat fragmentation is therefore essential for the conservation of biodiversity (Allendorf et al. 2012).
The expanding accessibility of genome sequencing technologies and analytical tools has greatly facilitated studies of functional connectivity and gene flow across a diverse array of rare, threatened, and ecologically important species (Manel and Holderegger 2013). However, inferring patterns of gene flow from genomic data remains a fundamental challenge in population genetics and landscape genetics (Cruickshank and Hahn 2014; Richardson et al. 2016; Beichman et al. 2018). The challenge stems largely from the similar genetic patterns that may be produced by multiple demographic and ecological processes (e.g., see Lawson et al., 2018; Meirmans, 2012). For instance, regions of a species’ range marked by relatively sharp transitions in genetic differentiation may occur when current gene flow is restricted by environmental gradients (Manthey and Moyle 2015; Weber et al. 2017), physical barriers (e.g., mountain ranges, rivers, or roads; Frantz et al., 2012), or poor habitat quality or habitat disturbance (Williams et al. 2003). Often, spatial patterns of genetic differentiation are correlated with landscape-level variables to infer putative ecological mechanisms shaping connectivity (i.e., landscape genetics; Manel et al. 2003). However, a central assumption of many correlative landscape genetics approaches is that contemporary patterns of genetic variation reflect a historical equilibrium of gene flow and genetic drift (Manel and Holderegger 2013), an assumption that may be seldom met in species with recently fragmented ranges (Whitlock and McCauley 1999). If the strength of gene flow or genetic drift has changed through time (e.g., recent dispersal barriers or admixture between previously isolated lineages), then traditional landscape genetic inferences may be confounded (Landguth et al. 2010; Epps and Keyghobad 2015). Understanding population history (i.e., demographic history) is therefore crucial for determining whether particular regions of a species’ range warrant focused management actions to increase connectivity (Epps and Keyghobad 2015).
Graham’s beardtongue (Penstemon grahamii D.D. Keck; Plantaginaceae) is a rare perennial forb endemic to the Uinta Basin of eastern Utah and western Colorado. Populations are patchily distributed across the oil shale-rich Green River formation, but the species is relatively common where it occurs (total population census estimate of 56,385 individuals across 9585 acres of occupied habitat; U.S. Fish and Wildlife Service 2020). P. grahamii is adapted to xeric environments (annual precipitation ~ 6–12 inches) and adult plants appear capable of going into dormancy during exceptionally dry conditions, which may facilitate its long-term persistence (U.S. Fish and Wildlife Service 2020). Within a year, temperatures can vary substantially across its range, with average summer highs reaching approximately 34 °C (~ 93°F) and average winter lows of − 14 °C (~ 7°F). P. grahamii seeds are primarily gravity- or wind-dispersed and pollinators include bees, wasps, and occasionally hummingbirds. Pollination is believed to be crucial to the reproductive success of P. grahamii, yet it remains unclear whether dispersal and pollination are sufficient for maintaining genetic variation across its range.
Penstemon grahamii faces numerous threats from energy development, mining, off-road vehicle use, and livestock disturbance. Energy development is particularly threatening, as half of known P. grahamii occurrences and 5 of 6 of its largest occurrences are on lands currently leased for oil and gas exploration (US Fish and Wildlife Service 2005). Individuals are also relatively long-lived (up to 10 years) and populations may have low recruitment (US Fish and Wildlife Service 2005), which naturally limits their ability to recover from disturbances that can persist across multiple generations. Given these factors, the federal protection status of P. grahamii has been the focus of substantial litigation, with proposals to list the species as endangered or threatened under the US Endangered Species Act in 1975, 1990, 2002, 2006, and 2013 (US Fish and Wildlife Service 2005, 2020). To inform ongoing conservation efforts, there is a pressing need to characterize genetic diversity and infer connectivity across the species’ range.
Here, we performed double-digest restriction site-associated DNA sequencing (ddRADseq) to investigate range-wide patterns of genetic diversity and functional connectivity in P. grahamii. First, to understand broad patterns of genetic structure reflecting historical gene flow across the range, we characterized the spatial distribution of genetic variation using principal component and population structure analyses. Then, to more precisely understand the historical demographic factors shaping these spatial patterns of genetic diversity and population structure we modeled the history of divergence, gene flow, and population size between P. grahamii lineages. Specifically, we were interested in testing whether P. grahamii lineages have experienced recent histories of isolation or secondary contact, which might influence the interpretation of recent gene flow based on population structure patterns. Our study provides valuable insights into functional connectivity of P. grahamii and highlights important and often overlooked challenges in elucidating the timing of gene flow from genomic data in species of conservation concern. Specifically, this research reinforces the idea that correct interpretation of contemporary genetic patterns first necessitates knowledge of historical demographic processes.
Methods
Sample collection, DNA isolation and library preparation
To obtain representative samples spanning the distribution of P. grahamii, we visited the 27 Element Occurrences (EOs) of P. grahamii defined by the U.S. Fish and Wildlife Service (U.S. Fish and Wildlife Service 2020) during the summer of 2019. Here, EOs are defined as occupied habitats separated from other occupied habitats by at least 2 km (km) (U.S. Fish and Wildlife Service 2020). Plants were found at 11 EOs across 22 individual sites and leaf tissues were sampled from 2 to 11 plants at each site (average 8.7 individuals per site, 192 individuals total; Supplementary Table 1) separated from one another by at least 5 m to avoid sampling close relatives.
We extracted genomic DNA using DNeasy 96 Plant Kits (Qiagen, Germantown, MD, USA) following the manufacturer’s protocol. DNAs were individually barcoded and processed into ddRAD libraries using EcoRI and MspI restriction enzymes (Peterson et al. 2012). Individual libraries were pooled and amplified using 18 polymerase chain reaction cycles. Amplified libraries were then size-selected using a Pippin Prep (Sage Science, Beverly, MA, USA) to isolate fragments ranging in length from 400 to 600 base pairs (bp). The final pooled library was sequenced on a HiSeq 4000 (Illumina, San Diego, CA, USA) at the University of Oregon’s Genomics and Cell Characterization Core Facility to generate single-end, 100 bp reads.
Genomic data preprocessing
Raw sequence data were processed and aligned using Stacks v2.41 (Rochette et al. 2019). We cleaned raw data by removing reads containing uncalled bases or with an average phred-scaled quality score below 22 within a 15 bp sliding window. We also trimmed the last 10 bp from high-quality reads due to declining quality scores. We determined the appropriate de novo assembly and filtering parameter settings in Stacks following Rochette & Catchen (2017). Briefly, we first processed a test data set while varying the parameters M (the maximum nucleotide distance allowed between stacks) and n (the number of mismatches allowed between loci during catalog construction) from 1 to 9 and while keeping M = n and setting m = 3 (minimum depth of coverage to create a stack). The test data set consisted of 12 samples from 12 sites (1 random sample per site; mean coverage 17.27 ×) that were chosen to evenly span the range (see Supplementary Table 1). Based on these tests, we processed the full data set with M = 5 and n = 5 because the total number of loci retained in 80% of individuals (r80) and the distribution of the number of single nucleotide polymorphisms (SNPs) per locus stabilized at these values (Supplementary Fig. 1). Next, we filtered out singletons, sites with heterozygosity > 0.75, and sites covered by fewer than 50% of individuals. Finally, we restricted our data set to include only 1 SNP per locus to minimize the number of non-independent SNPs (i.e., reduce linkage in the data set).
Population structure
We inferred genetic structure patterns using a principal components analysis (PCA) in EIGENSOFT v6.1.4 (Patterson et al. 2006). In addition, we performed population cluster analyses in Structure (Pritchard et al. 2000) and tested K values (representing the number of population clusters) from 1 to 8, performing 5 iterations under each K value. For each iteration, we performed 100,000 MCMC repetitions following a burn-in of 20,000 repetitions assuming a diploid model (other parameters were set to default values). We used the program Structure Harvester v0.6.94 (Earl and VonHoldt 2012) to determine the optimal K value using the Evanno method (Evanno et al. 2005). We calculated Weir and Cockerham’s FST between the Structure-defined genetic clusters using vcftools (Danecek et al. 2011).
To examine associations between the spatial distribution of genetic variation and geography, we performed a Procrustes analysis (Wang et al. 2010). In a population genetic context, a Procrustes analysis transforms (i.e., rotates and scales) a two-dimensional principal component (PC) plot of genetic data to match a geographic map of sampling locations, with the goal of minimizing the overall distance between the two data sets (Wang et al. 2010). When genetic relatedness is strongly correlated with geographic proximity (i.e., isolation-by-distance; IBD; Wright 1943), genetic PC plots are expected to mirror the geographic sampling map. As a result, Procrustes analyses can highlight deviations from an IBD pattern that arise from biological or historical phenomena (e.g., Knowles et al. 2016; Wang et al. 2012). A Procrustes analysis was performed with the PC axes generated in EIGENSOFT using the ‘protest’ function of the VEGAN package (Oksanen et al. 2013) in R. The significance of the association statistic (t0) between the genetic PC values and geography was evaluated based on 10,000 permutations, where geographic locations were randomly permuted across the different sampling localities (note that all individuals from the same locality were assigned to a single geographic location in the permuted data set, such that observed levels of population structure were maintained).
Effective migration and diversity surfaces
We visualized spatially explicit patterns of genetic diversity and population structure using the program EEMS (Estimated Effective Migration Surfaces; Petkova et al. 2016). EEMS models the relationship between genetic variation and geography assuming stepping-stone migration between a predefined number of demes (i.e., occupied sites) arranged evenly in a grid across a species’ range. Landscape surface models of ‘effective diversity’ (i.e., relative genetic dissimilarity within demes) and ‘effective migration’ (i.e., relative genetic dissimilarity between demes) are fit to empirical genetic data, with sampled sites assigned to the closest deme on the landscape grid. Good models are those with a high correlation between observed and expected genetic dissimilarity both within and between demes. The effective migration surface allows the identification of specific geographic regions that deviate from an IBD pattern of genetic differentiation (i.e., with higher or lower than expected decay in genetic similarity across space).
We performed EEMS analyses assuming either 20, 50, or 100 demes across the P. grahamii range. These deme values represent a biologically realistic number of occupied sites based on the current estimated number of EOs (n = 27; U.S. Fish and Wildlife Service 2020) while allowing higher values due to the potential for incomplete survey information across the species’ range. For each deme value, we performed 3 independent runs and checked results for consistency between runs. We used default hyper-parameter values and tuned the proposal variances such that proposals were accepted approximately 30% of the time. We ran EEMS for 2 million iterations with a burn-in of 1 million iterations and thinning iteration of 9999.
Inferring admixture and demographic history
We used the program ∂a∂i (Gutenkunst et al. 2009) to estimate the timing of population divergence (t), ancestral effective population sizes (Nanc), contemporary effective population sizes (N), and the timing and level of gene flow between geographically adjacent genetic clusters identified with Structure (W and C clusters, C and SE clusters, and SE and NE clusters; see Results). To represent the C and SE genetic clusters, we used individuals from a subset of sites that were geographically closest to the paired cluster being analyzed. We inferred demographic parameters using a folded site frequency spectrum (SFS; i.e., major/minor allele calls rather than derived/ancestral) due to the lack of a closely related reference genome. We scaled the t and m parameters (i.e., reported in coalescent units by ∂a∂i) by θ, assuming the tomato neutral mutation rate (μ = 1e–8; Lin et al., 2014) and a 121:1 ratio of callable sites:SNPs (based on output from Stacks).
For each pair of genetic clusters, we tested four alternative demographic models: (1) no migration, (2) isolation-with-migration (IM), (3) recent admixture, and (4) recent isolation (see Fig. 2A). The IM model includes a symmetrical migration rate (m; migrants per generation) between populations immediately following divergence t generations ago. Under the admixture model, populations are initially isolated for t1 generations and then experience symmetrical migration starting t2 generations in the past. Finally, the recent isolation model includes symmetrical migration between populations for t1 generations following divergence, after which migration ceases t2 generations ago. For each model, we performed 20 independent runs starting with parameter values sampled randomly across a uniform distribution (0.01 < ν < 100, 0 < 2Nanct < 20, 0 < 2Nancm < 5; here, ν is the relative change in population size compared to Nanc). This modeling strategy was designed to test among simple alternative scenarios that may approximate the actual population histories underlying contemporary patterns of genetic differentiation and not to elucidate the full, complex evolutionary histories of these populations. Larger genomic data sets may be able to discern among more complex (i.e. parameter-rich) models, but this is beyond the scope of our study.
We determined which of the four maximum likelihood demographic models produced the best overall fit to the data using a composite-likelihood ratio test with the Godambe Information Matrix (GIM; Coffman et al. 2016) and Akaike Information Criterion (AIC) scores. Confidence intervals (CI) for each parameter were estimated with the Godambe Information Matrix with 100 bootstrap data sets comprising 30% of SNPs randomly selected from the full data set. We validated the final model by comparing the predicted SFS to the observed SFS for each population.
Results
Population genetic structure and diversity
Across 192 P. grahamii individuals, we obtained 7805 SNPs from a total of 941,633 sequenced sites (0.83% of sites were polymorphic; mean sequence coverage of 17.3 ± 1.8 × standard deviation). Nucleotide diversity was 0.00122 ± 0.00002 standard error and FIS was 0.0013 ± 0.02885 standard error across the entire data set (see Supplementary Table 1 for summary statistics for individual sampling sites).
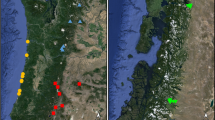
A range-wide PCA and Procrustes analysis revealed genetic variation largely follows an IBD pattern, with individuals approximately clustering according to their geographic sampling coordinates (Fig. 1A, Supplementary Fig. 2B). Procrustes transformations uncovered a strong and highly significant correlation (t0 = 0.86; p = 0.0001) between individual’s coordinates in PC space and sampling locations in geographic space. Structure analyses revealed that a K = 4 model provided the best fit to the data (Supplementary Fig. 3). Under K = 4, we found genetic clusters corresponding to the 3 western-most sites (hereafter, the W cluster), 9 central sites (C cluster), 8 southeastern sites (SE cluster), and 2 northeastern sites (NE cluster; see Fig. 1B). Estimates of genome-wide pairwise FST between genetically defined clusters were relatively low (W and C clusters FST = 0.039; C and SE clusters FST = 0.016; SE-NE clusters FST = 0.026, W and NE clusters FST = 0.06). The 95% empirical quantiles for FST estimated across individual RAD loci were − 0.024–0.0272 for W and C cluster, − 0.015–0.112 for C and SE clusters, − 0.052–0.289 for SE and NE clusters, and − 0.050–0.394 for W and NE clusters.
A A map of Penstemon grahamii sampling localities (white diamonds) in Utah and Colorado. Individual plants are represented by colored points and are positioned relative to one another in two principal component axes based on a Procrustes analysis (black lines connect individuals to their sampling locality). Under a perfect isolation-by-distance (IBD) scenario, individual points are expected to be placed on top of their sampling locality. Here, our results depict a highly significant IBD relationship across the P. grahamii range (t0 = 0.86, p = 0.0001). B Spatial visualization of the optimal K = 4 Structure results. The proportion of each color in each pie represents the average proportion of ancestry assigned to that genetic cluster at each locality. The arrows and numbers indicate pairwise FST values between the indicated clusters. C Effective migration surface estimated in EEMS using 50 demes. Blue values indicate regions of relatively high effective migration (m) while orange values indicate regions of relatively low effective migration (here, log10(m) = 1 indicates tenfold faster migration compared to the average across the range; Petkova et al., 2016). Circles indicate occupied deme localities with the size of the circle corresponding to the number of samples
We found evidence of mixed ancestry across sites, particularly those along the approximate edges of each cluster (Fig. 1B), which may reflect continuous population structure (i.e., IBD) or admixture between discrete genetic clusters. It is well established that the presence of IBD can confound clustering algorithms, leading to false inferences of hierarchical population structure, and vice versa (Meirmans 2012; Lawson et al. 2018). Acknowledging this uncertainty, we also present results for K = 2 and K = 3 (Supplementary Fig. 2C) to demonstrate general spatial genetic patterns that emerged from the data. Under K = 2, individuals from the three western sites formed a distinct cluster relative to the individuals across the remainder of the range, which was expected based on similar patterns in the PCA. Under K = 3, we found the main genetic cluster under K = 2 breaks into two clusters, resulting in a western, central, and eastern cluster.
Range-wide effective diversity and migration
EEMS runs produced strong correlations between observed and expected genetic dissimilarity both within and between demes, indicating good model fit (Petkova et al. 2016; Supplementary Fig. 4). Under the 20 deme model, only 6 demes were represented by empirical data, resulting in poor spatial resolution of effective diversity and migration (Supplementary Fig. 4). We therefore focus on the results for the 50 and 100 deme models. Across both deme values, effective diversity surfaces revealed relatively low genetic diversity towards the eastern and western extents of the range (~ 10–15% lower than average diversity under the 50 deme model; Supplementary Fig. 4). Reduced variation at the range edges is unsurprising as populations along range margins are often predicted to occur at low densities, resulting in lower levels of genetic diversity (Antonovics 1976; Kirkpatrick and Barton 1997; Pfennig et al. 2016).
We found evidence of average to somewhat low effective migration across the western portion of the range (spanning a ~ 50 km sampling gap; Fig. 1C, Supplementary Fig. 4). This pattern is generally suggestive of IBD and potentially indicates the presence of suitable habitat across our sampling gap. However, we uncovered several regions of strongly reduced effective migration, including one across the eastern part of the range and one in the center of the range (Fig. 1C and Supplementary Fig. 4). This pattern was particularly striking under the 50 deme model, where effective migration was ~ 5–30-fold lower than average in these regions, indicating a significant deviation from a homogenous IBD pattern.
The history of population divergence and gene flow
Demographic modeling of Structure-defined genetic clusters (K = 4) revealed relatively high rates of historical gene flow across the range. Across the western portion of the range (W and C clusters), we found strong support for an IM model over a model without migration (AIC p-value = 1.3e–62). Under the IM model, populations diverged ~ 72 thousand generations ago and experienced a gene flow rate (m) of 1.13 migrants/generation (Table 1). Moreover, we uncovered relatively large effective population sizes for each cluster (W cluster Ne = 24,905, C cluster Ne = 48,104, Nanc = 15,824). Similarly, we found strong support for an IM model between the SE and NE clusters over a model with no migration (AIC p-value = 1.1e–30), with a divergence time of ~ 115 thousand generations, 1.84 migrants/generation, and large effective population sizes (SE cluster Ne = 34,874, NE cluster Ne = 25,886, Nanc = 9848). Although an IM model was the best supported model based on AIC scores for both W-C and C-SE population pairs, we were unable to rule out an admixture model (for both pairs) or a recent isolation model (for W and C clusters), as these more complex models produced a similar overall fit to the data (see Supplementary Fig. 5). Between the C and SE clusters, however, we found significant support for an admixture model over an IM model (GIM p-value = 1.28e–6; AIC p-value = 0.002). Here, we estimated that populations diverged under isolation ~ 40 thousand generations ago (although confidence intervals for the initial divergence date are large (standard error = 24,411 generations); Table 1) and ~ 3388 generations ago gene flow between populations began occurring at a rate of 3.23 migrants/generation. Estimates of effective population sizes under this model were similarly large (C cluster Ne = 29,802, SE cluster Ne = 13,468, Nanc = 8969).
To make inferences into relative rates of gene flow across P. grahamii’s distribution, we standardized our estimates of m between clusters by the average geographic distance between sites used to represent each cluster in the analysis. Thus, our standardized estimates of gene flow are 56.3 migrants/gen/km between W and C clusters, 49.1 migrants/gen/km between C and SE clusters, and 46.6 migrants/gen/km between SE and NE clusters.
Discussion
The maintenance of functional connectivity between populations of rare species experiencing habitat fragmentation is essential for the conservation of biodiversity (Allendorf et al. 2012). As such, characterizing population structure and gene flow within threatened and endangered species is necessary to identify appropriate management units (Palsbøll et al. 2007) and prescribe adequate recovery plans (Allendorf et al. 2012). However, as we demonstrate through our investigation of spatial genetic patterns in P. grahamii, correct interpretation of contemporary genetic patterns may also necessitate the inclusion of a historical perspective to best inform management.
Population structure and demographic history of P. grahamii
Inferences of population structure within P. grahamii offered potentially conflicting interpretations regarding the extent of functional connectivity across contemporary populations. A genome-wide PCA and Procrustes analysis uncovered a general IBD pattern (Fig. 1A, Supplementary Fig. 2B), which arises when local demes are connected to adjacent demes through migration (Wright 1943). Likewise, Structure revealed a generally continuous pattern of population structure across the range, potentially indicative of IBD or admixture between adjacent, hierarchically defined populations. Notably, however, we observed striking shifts in genetic ancestry between the inferred C (green) and SE (pink) clusters and the SE and NE (blue) clusters across a small spatial scale, which could indicate historically reduced gene flow in these regions. In support of this interpretation, effective migration surfaces indicated regions marked by relatively strong genetic differentiation (~ 5–30-fold lower effective migration than average) near the central and eastern portions of the range (Fig. 1C).
To gain more detailed insights into the genetic patterns that influence the interpretation of functional connectivity across P. grahamii populations, we fit empirical genetic data to a suite of alternative demographic models allowing for different histories of gene flow between adjacent population pairs (Fig. 2A). Although we were unable to conclusively determine the precise historical model (e.g., IM versus admixture) for two population pairs (W-C and SE-NE), we can rule out models without gene flow for all populations. For W-C and SE-NE comparisons, continuous migration following population divergence was the most likely model, consistent with population genetic analyses described above. Prior to this study, it was unclear whether suitable P. grahamii habitat exists between the W and C clusters (a ~ 50 km gap; Fig. 1) because permitting restrictions prevented surveys. Our results indicate high historical functional connectivity across the western portion of the range, suggesting suitable habitat is likely present and occupied. This is consistent with previous ecological modeling work in P. grahamii showing relatively continuous predicted suitable habitat across this portion of the range (Decker et al. 2006). In contrast to the IM model between populations within the eastern and western portions of the species’ range, demographic modeling suggests that gene flow commenced more recently at the range center between the C and SE clusters (Table 1). Recent admixture would explain the conflicting patterns revealed by population structure analyses—specifically, that low effective migration at the range center could reflect historical isolation rather than ongoing restricted gene flow—and provide guidance for appropriate management actions, which we discuss below.
A Illustration of alternative demographic models tested in ∂a∂i (Gutenkunst et al. 2009). Parameters include ancestral effective population size (Nanc), contemporary effective population sizes (N1 and N2), migration rate (m, migrants per generation), the timing of population divergence (t), and the timing of changes in migration rates (t2). B Illustration of best fit demographic modeling results for three pairwise population comparisons (W-C, C-SE, and SE-NE). The thickness of lines indicates relative differences in population sizes and migration rates
Management implications
Recent anthropogenic-mediated changes in the western US (e.g., habitat destruction, drought, wildfires, grazing, and the spread of invasive species) have been linked to range fragmentation and reduced connectivity in a number of native plant and animal species (Hargis et al. 1999; Theobald et al. 2011; Pilliod et al. 2015; Warren et al. 2016). Similar to the challenges faced by other rare species, small and fragmented populations of P. grahamii are likely vulnerable to habitat destruction and modification from oil and gas development, livestock, or other human-mediated alterations (US Fish and Wildlife Service 2005). Yet, it remains unclear how to design management efforts to mitigate these influences. By assessing both contemporary patterns of genetic diversity and differentiation and historical population dynamics, we can illuminate the processes affecting connectivity and develop more informed species recovery plans.
Our study suggests that, despite range fragmentation, P. grahamii can be treated as a single management unit because the low genetic differentiation across the species’ range (pairwise FST ≈ 0.02–0.06) appears to be the result of high levels of historical gene flow (at least since ~ 3400 generations ago; Table 1). Moreover, large effective population size estimates (similar to population census size estimates) suggest a low risk of inbreeding depression and the potential for local adaptation within populations. Therefore, conservation efforts may be optimized by maintaining or enhancing opportunities for local dispersal through habitat preservation wherever suitable habitat is less abundant or occupied sites are geographically distant, as gene flow between similar environments will help maintain existing genetic diversity while not swamping potential patterns of local adaptation.
Two geographic regions across the P. grahamii range may disproportionately benefit from management efforts. Firstly, we find the lowest standardized rates of historical gene flow (46.6 migrants/gen/km) across the eastern range edge where genetic diversity is also relatively low. This portion of the range has experienced the most intensive oil and gas development (US Fish and Wildlife Service 2005). Focused efforts to preserve habitat and facilitate gene flow may help maintain these low diversity edge populations, which are often essential populations for the long-term persistence of species under changing environments (Rehm et al. 2015). We also find that the range center appears to be a recent contact zone between eastern and western lineages. Contingent on the preservation of habitat, the range center is likely an important corridor for the transfer of unique genetic variation between previously isolated lineages, which could support adaptive responses to rapidly changing environments (e.g., Hamilton and Miller 2016; Jones et al. 2018; Pardo-Diaz et al. 2012; Song et al. 2011). Future studies with expanded genomic and individual sampling may be able to assess more recent levels of gene flow across different portions of the range, which would further improve our understanding of functional connectivity and help guide management efforts.
On the use of demographic models in conservation
Although it is widely recognized that historical processes can produce non-equilibrium spatial patterns of genetic variation (Landguth et al. 2010; Manel and Holderegger 2013; Epps and Keyghobad 2015; Aitken and Bemmels 2016), many landscape genetic studies still fail to consider historical perspectives when analyzing current patterns of population structure (e.g., Aavik et al. 2014; Milanesi et al. 2017; Parks et al. 2015). Our study illustrates the potential danger of neglecting historical demography when investigating functional connectivity in rare species and the value of demographic models for guiding conservation decision-making.
The assumption of migration-drift equilibrium in P. grahamii could lead to the erroneous conclusion that the range center experiences significantly reduced gene flow relative to other parts of the range. Instead, demographic modeling reveals the striking shift in population ancestry and low effective migration in this region is likely the result of recent secondary contact between previously isolated lineages, with a rate of gene flow (49.1 migrants/generation/km) similar to other population pairs. Importantly, if our demographic model is accurate, parameter estimates indicate populations at the range center are likely not in migration-drift equilibrium. Thus, simple correlations between spatial patterns of genetic variation and landscape-level variables would likely produce misleading inferences into ecological factors shaping gene flow. In general, the problem of inferring spatial patterns of gene flow in non-equilibrium populations may be alleviated in rare or geographically restricted species, as small populations reach migration-drift equilibrium more quickly than large populations (Whitlock and McCauley 1999). However, as shown in this and other studies (e.g., Barrett and Kohn 1991; Ellstrand and Elam 1993; Jones et al. 2021a), it is not uncommon for endemic species to be locally abundant or for rare species to have unexpectedly high genetic diversity. We therefore recommend that demographic modeling be performed in conjunction with correlative landscape genetic analyses to determine whether populations likely meet the assumption of migration-drift equilibrium.
Beyond the importance of accounting for historical processes in landscape genetic studies of functional connectivity, demographic models themselves have important value for conservation. In particular, the estimation of effective population size and migration rates are essential to the development of conservation strategies (Mills and Allendorf 1996; Leitwein et al. 2020). Understanding effective population sizes is crucial for guiding management actions towards the populations that are most susceptible to inbreeding depression and the accumulation of deleterious mutations (Lynch et al. 1995; Charlesworth and Willis 2009; Harmon and Braude 2010). For instance, an effective population size of at least 50 is expected to reduce the chances of inbreeding depression in the short-term, while an effective population size of at least 500 is required to minimize the loss of genetic diversity due to drift (Harmon and Braude 2010). Moreover, inferences of migration rates between populations may aid in determining where efforts to enhance connectivity should be focused to preserve genetic diversity (e.g., based on the one-migrant-per-generation rule; Mills and Allendorf 1996). Beyond inferring rates of gene flow, understanding historical changes in gene flow can also provide important context for conservation decision-making. For instance, here, the identification of a putative region of admixture in P. grahamii may be important for developing management strategies to facilitate the exchange and maintenance of genetic variation between previously isolated lineages.
Although estimating demographic history has important applications for conservation, we urge caution when applying demographic modeling to rare species due to the known limitations of the analytical framework. For instance, inferring recent historical events (e.g., hundreds of generations in the past) using SFS-based demographic modeling is difficult or impossible without very large sample sizes (Robinson et al. 2014; Beichman et al. 2018). If P. grahamii experienced recent population declines or fragmentation, then current levels of gene flow may be lower than our demographic model estimates. Demographic estimation can also be strongly skewed if genetic loci experience linked selection (Ewing and Jensen 2016; Johri et al. 2020), which is usually impossible to infer in non-model species. Moreover, distinct demographic models can produce similar fits to the same genetic data set, as we have observed in this study (Supplementary Fig. 5), and thus testing, comparing, and validating alternative historical models is essential. Throughout, we avoided interpreting our modeling results as reflecting the precise history of these populations or contemporary demographic processes. Rather demographic modeling gives an indication of historical averages of gene flow, which likely fluctuate over time in magnitude and direction. With a clear understanding of the limitations and assumptions inherent to these methods, the inclusion of a historical perspective is critical to correctly interpret genetic patterns and thus prescribe management recommendations for rare, threatened, or endangered species.
Data availability
Data generated from this study are available at the USGS ScienceBase-Catalog (Jones et al. 2021b).
Code availability
Code generated for this study are available at the USGS ScienceBase-Catalog (Jones et al. 2021b).
References
Aavik T, Holderegger R, Bolliger J (2014) The structural and functional connectivity of the grassland plant Lychnis flos-cuculi. Heredity 112:471–478
Aitken SN, Bemmels JB (2016) Time to get moving: assisted gene flow of forest trees. Evol Appl 9:271–290. https://doi.org/10.1111/eva.12293
Allendorf FW, Luikart G, Aitken SN (2012) Conservation and the genetics of populations, 2nd edn. Wiley-Blackwell, Malden
Antonovics J (1976) The nature of limits to natural selection. Ann Missouri Bot Gard 63:224–247
Barrett SCH, Kohn JR (1991) Genetic and evolutionary consequences of small population size in plants: implications for conservation. In: Falk DE, Holsinger KE (eds) Genetics and conservation of rare plants. Oxford University Press, Oxford, pp 3–30
Beichman AC, Huerta-Sanchez E, Lohmueller KE (2018) Using genomic data to infer historic population dynamics of nonmodel organisms. Annu Rev Ecol Evol Syst 49:433–456. https://doi.org/10.1146/annurev-ecolsys-110617
Charlesworth D, Willis JH (2009) The genetics of inbreeding depression. Nat Rev Genet 10:783–796
Coffman AJ, Hsieh PH, Gravel S, Gutenkunst RN (2016) Computationally efficient composite likelihood statistics for demographic inference. Mol Biol Evol 33:591–593. https://doi.org/10.1093/molbev/msv255
Crooks KR, Sanjayan M (2006) Connectivity conservation: maintaining connections for nature. Connectivity conservation. Cambridge University Press, Cambridge
Cruickshank TE, Hahn MW (2014) Reanalysis suggests that genomic islands of speciation are due to reduced diversity, not reduced gene flow. Mol Ecol 23:3133–3157. https://doi.org/10.1111/mec.12796
Danecek P, Auton A, Abecasis G et al (2011) The variant call format and VCFtools. Bioinformatics 27:2156–2158. https://doi.org/10.1093/bioinformatics/btr330
Decker K, Lavender A, Handwerk J, Anderson DG (2006) Modeling the potential distribution of three endemic plants of the Northern Piceance and Uinta Basins. Colorado State University, Fort Collins
Earl DA, VonHoldt BM (2012) Structure harvester: a website and program for visualizing structure output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Ellstrand NC, Elam DR (1993) Population genetic consequences of small population size: implications for plant conservation. Annu Rev Ecol Syst 24:217–259. https://doi.org/10.1146/annurev.es.24.110193.001245
Epps CW, Keyghobad N (2015) Landscape genetics in a changing world: disentangling historical and contemporary influences and inferring change. Mol Ecol 24:6021–6040
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14:2611–2620
Ewing GB, Jensen JD (2016) The consequences of not accounting for background selection in demographic inference. Mol Ecol 25:135–141. https://doi.org/10.1111/mec.13390
Fisher RA (1930) The genetic theory of natural selection. Clarendon Press, London
Frantz AC, Bertouille S, Eloy MC et al (2012) Comparative landscape genetic analyses show a Belgian motorway to be a gene flow barrier for red deer (Cervus elaphus), but not wild boars (Sus scrofa). Mol Ecol 21:3445–3457
Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD (2009) Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet 5:e1000695. https://doi.org/10.1371/journal.pgen.1000695
Hamilton JA, Miller JM (2016) Adaptive introgression as a resource for management and genetic conservation in a changing climate. Conserv Biol 30:33–41. https://doi.org/10.1111/cobi.12574
Hargis CD, Bissonette JA, Turner DL (1999) The influence of forest fragmentation and landscape pattern on American martens. J Appl Ecol 36:157–172
Harmon LJ, Braude S (2010) Conservation of small populations: effective population sizes, inbreeding, and the 50/500 rule. An introduction to methods and models in ecology, evolution, and conservation biology. Princeton University Press, New Jersy, pp 125–138
Hughes AR, Inouye BD, Johnson MTJ et al (2008) Ecological consequences of genetic diversity. Ecol Lett 11:609–623. https://doi.org/10.1111/j.1461-0248.2008.01179.x
Johri P, Charlesworth B, Jensen JD (2020) Towards an evolutionarily appropriate null model: jointly inferring demography and purifying selection. Genetics 215:173–192
Jones MR, Mills LS, Alves PC et al (2018) Adaptive introgression underlies polymorphic seasonal camouflage in snowshoe hares. Science 360:1355–1358. https://doi.org/10.1126/science.aar5273
Jones MR, Winkler DE, Massatti R (2021a) The demographic and ecological factors shaping diversification among rare Astragalus species. Divers Distrib. https://doi.org/10.1111/ddi.13288
Jones MR, Winkler DE, Massatti R (2021b) Penstemon grahamii genetic data from a dryland region of the western United States: U.S. Geological Survey data release. https://doi.org/10.5066/P9VRF7AR
Kirkpatrick M, Barton NH (1997) Evolution of a species’ range. Am Nat 150:1–23. https://doi.org/10.1086/286054
Knowles LL, Massatti R, He Q et al (2016) Quantifying the similarity between genes and geography across Alaska’s alpine small mammals. J Biogeogr 43:1464–1476
Landguth EL, Cushman SA, Schwartz MK et al (2010) Quantifying the lag time to detect barriers in landscape genetics. Mol Ecol 19:4179–4191
Lawson DJ, van Dorp L, Falush D (2018) A tutorial on how not to over-interpret structure and admixture bar plots. Nat Commun 9:3258
Leitwein M, Duranton M, Rougemont Q et al (2020) Using haplotype information for conservation genomics. Trends Ecol Evol 35:245–258
Lin T, Zhu G, Zhang J et al (2014) Genomic analyses provide insights into the history of tomato breeding. Nat Genet 46:1220–1226
Lynch M, Conery J, Burger R (1995) Mutation accumulation and the extinction of small populations. Am Nat 146:489–518. https://doi.org/10.1086/285812
Manel S, Holderegger R (2013) Ten years of landscape genetics. Trends Ecol Evol 28:614–621
Manel S, Schwartz MK, Luikart G, Taberlet P (2003) Landscape genetics: combining landscape ecology and population genetics. Trends Ecol Evol 18:189–197
Manthey JD, Moyle RG (2015) Isolation by environment in White-breasted Nuthatches (Sitta carolinensis) of the Madrean Archipelago sky islands: a landscape genomics approach. Mol Ecol 24:3628–3638
Mantyka-Pringle CS, Martin TG, Rhodes JR (2012) Interactions between climate and habitat loss effects on biodiversity: a systematic review and meta-analysis. Glob Chang Biol 18:1239–1252
Meirmans PG (2012) The trouble with isolation by distance. Mol Ecol 21:2839–2846
Milanesi P, Holderegger R, Bollmann K et al (2017) Three-dimensional habitat structure and landscape genetics: a step forward in estimating functional connectivity. Ecology 98:393–402
Mills LS, Allendorf FW (1996) The one-migrant-per-generation rule in conservation and management. Conserv Biol 10:1509–1518. https://doi.org/10.1046/j.1523-1739.1996.10061509.x
Oksanen J, Blanchet FG, Kindt R et al (2013) Package Vegan. Community ecology package version 2.0–10
Palsbøll PJ, Bérubé M, Allendorf FW (2007) Identification of management units using population genetic data. Trends Ecol Evol 22:11–16
Pardo-Diaz C, Salazar C, Baxter SW et al (2012) Adaptive introgression across species boundaries in Heliconius butterflies. PLoS Genet 8:e1002752. https://doi.org/10.1371/journal.pgen.1002752
Parks LC, Wallin DO, Cushman SA, McRae BH (2015) Landscape-level analysis of mountain goat population connectivity in Washington and southern British Columbia. Conserv Genet 16:1995
Patterson N, Price AL, Reich D (2006) Population structure and eigenanalysis. PLoS Genet 2:e190. https://doi.org/10.1371/journal.pgen.0020190
Peterson BK, Weber JN, Kay EH et al (2012) Double digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS ONE 7:e37135. https://doi.org/10.1371/journal.pone.0037135
Petkova D, Novembre J, Stephens M (2016) Visualizing spatial population structure with estimated effective migration surfaces. Nat Genet 48:94–100. https://doi.org/10.1038/ng.3464
Pfennig KS, Kelly AL, Pierce AA (2016) Hybridization as a facilitator of species range expansion. Proc R Soc B Biol Sci 283:20161329. https://doi.org/10.1098/rspb.2016.1329
Pilliod DS, Arkle RS, Robertson JM et al (2015) Effects of changing climate on aquatic habitat and connectivity for remnant populations of a wide-ranging frog species in an arid landscape. Ecol Evol 5:3979–3994
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Rehm EM, Olivas P, Stroud J, Feeley KJ (2015) Losing your edge: climate change and the conservation value of range-edge populations. Ecol Evol 5:4315–4326
Richardson JL, Brady SP, Wang IJ, Spear SF (2016) Navigating the pitfalls and promise of landscape genetics. Mol Ecol 25:849–863
Robinson JD, Coffman AJ, Hickerson MJ, Gutenkunst RN (2014) Sampling strategies for frequency spectrum-based population genomic inference. BMC Evol Biol 14:254. https://doi.org/10.1186/s12862-014-0254-4
Rochette NC, Catchen JM (2017) Deriving genotypes from RAD-seq short-read data using stacks. Nat Protoc 12:2640–2659. https://doi.org/10.1038/nprot.2017.123
Rochette NC, Rivera-Colón AG, Catchen JM (2019) Stacks 2: analytical methods for paired-end sequencing improve RADseq-based population genomics. Mol Ecol 28:4737–4754. https://doi.org/10.1111/mec.15253
Sgrò CM, Lowe AJ, Hoffmann AA (2011) Building evolutionary resilience for conserving biodiversity under climate change. Evol Appl 4:326–337. https://doi.org/10.1111/j.1752-4571.2010.00157.x
Song Y, Endepols S, Klemann N et al (2011) Adaptive introgression of anticoagulant rodent poison resistance by hybridization between old world mice. Curr Biol 21:1296–1301. https://doi.org/10.1016/j.cub.2011.06.043
Theobald DM, Crooks KR, Norman JB (2011) Assessing effects of land use on landscape connectivity: loss and fragmentation of western US forests. Ecol Appl 21:2445–2458
Travis JMJ (2004) Climate change and habitat destruction: a deadly anthropogenic cocktail. Proc R Soc B Biol Sci 270:467–473
US Fish and Wildlife Service (2005) US fish and wildlife service species assessment and listing priority assignment form: Penstemon grahamii, pp 9
U.S. Fish and Wildlife Service (2020) Final Graham’s beardtongue (Penstemon grahamii) and White River beardtongue (P. scariosus var. albifluvis): Biological status report of current condition and recommended avoidance buffer and surface disturbance caps. Utah Field Office, Ecological Services, U.S. Fish and Wildlife Service, West Valley City, Utah. December 21, 2020. pp 112 + Appendices.
Wang C, Szpiech ZA, Degnan JH et al (2010) Comparing spatial maps of human population-genetic variation using procrustes analysis. Stat Appl Genet Mol Biol 9:13
Wang C, Zöllner S, Rosenberg NA (2012) A quantitative comparison of the similarity between genes and geography in worldwide human populations. PLOS Genet 8:e1002886
Warren MJ, Wallin DO, Beausoleil RA, Warheit KI (2016) Forest cover mediates genetic connectivity of northwestern cougars. Conserv Genet 17:1011–1024
Weber JN, Bradburd GS, Stuart YB et al (2017) Partitioning the effects of isolation by distance, environment, and physical barriers on genomic divergence between parapatric threespine stickleback. Evolution 71:342–356
Whitlock MC, McCauley DE (1999) Indirect measures of gene flow and migration: FST≠1/(4Nm+1). Heredity 82:117–125
Williams BL, Brawn JD, Paige KN (2003) Landscape scale genetic effects of habitat fragmentation on a high gene flow species: Speyeria idalia (Nymphalidae). Mol Ecol 12:11–20
Wright S (1931) Evolution in Mendelian populations. Genetics 16:97–159
Wright S (1943) Isolation by distance. Genetics 28:114–138
Acknowledgements
We would like to thank Shannon Lencioni for assistance with field collections. We thank Joseph Moore for analytical assistance. We thank Gery Allan, Lela Andrews, and Erica Sukovich at Northern Arizona University’s Environmental Genetics and Genomics Laboratory for expert support with laboratory procedures. We would also like to thank Jennifer Lewinsohn, Rita Reisor, and the U.S. Geological Survey Ecosystem Mission Area for providing the opportunity for this research. Finally, we wish to thank two anonymous reviewers for their helpful comments and suggestions on earlier drafts of this manuscript. Any use of trade, product or firm names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
Funding
United States Geological Survey.
Author information
Authors and Affiliations
Contributions
DEW and RM performed biological sampling; MRJ, DEW, and RM designed research; MRJ performed data analyses; MRJ wrote the paper with contributions from both authors.
Corresponding author
Ethics declarations
Conflicts of interest
The authors declared that they have no conflict of interest.
Consent to participate
Consent granted.
Consent for publication
Consent granted.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jones, M.R., Winkler, D.E. & Massatti, R. Demographic modeling informs functional connectivity and management interventions in Graham’s beardtongue. Conserv Genet 22, 993–1003 (2021). https://doi.org/10.1007/s10592-021-01392-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-021-01392-9