Abstract
Establishment of adequate conservation areas represents a challenging but crucial task in the conservation of genetic diversity and biological variability. Anthropogenic pressures on marine ecosystems and organisms are steadily increasing. Whether and to what extent these pressures influence marine genetic biodiversity is only starting to be revealed. Using 16S rRNA gene amplicon sequencing, we analysed the microbial community structure of 33 individuals of the habitat-forming glass sponge Vazella pourtalesii, as well as reference seawater, sediment, and biofilm samples. We assessed how two anthropogenic impacts, i.e. habitat destruction by trawling and artificial substrate provision (moorings made of composite plastic), correspond with in situ V. pourtalesii microbiome variability. In addition, we evaluated the role of two bottom fishery closures in preserving sponge-associated microbial diversity on the Scotian Shelf, Canada. Our results illustrate that V. pourtalesii sponges collected from protected sites within fishery closures contained distinct and taxonomically largely novel microbial communities. At the trawled site we recorded significant quantitative differences in distinct microbial phyla, such as a reduction in Nitrospinae in the four sponges from this site and the environmental references. Individuals of V. pourtalesii growing on the mooring were significantly enriched in Bacteroidetes, Verrucomicrobia and Cyanobacteria in comparison to sponge individuals growing on the natural seabed. Due to a concomitant enrichment of these taxa in the mooring biofilm, we propose that biofilms on artificial substrates may ‘prime’ sponge-associated microbial communities when small sponges settle on such substrates. These observations likely have relevant management implications when considering the increase of artificial substrates in the marine environment, e.g., marine litter, off-shore wind parks, and petroleum platforms.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Glass sponges (Hexactinellida) are extraordinary animals with a skeleton made of silicon dioxide and a unique histology which is distinct from all other known sponge classes (Leys et al. 2007). The greatest taxonomic diversity of glass sponges is found between 300 and 600 m depth, with only a few populations occurring in shallow (euphotic) waters (Leys et al. 2007). Hexactinellids are among the most ancient metazoans with an estimated origin of 800 million years ago as determined by molecular clocks (Leys et al. 2007), and therefore represent promising candidates for examining evolutionary ancient biological mechanisms and relationships such as sponge-microbe interactions (Pita et al. 2018). The microbial communities associated with sponges can be very diverse, with more than 63 phyla reported previously (Thomas et al. 2016; Moitinho-Silva et al. 2017). In order to gain a holistic understanding of multicellular organisms, also their internal symbiotic associations should be considered (McFall-Ngai et al. 2013). Along similar lines, the term ‘holobiont’ (syn. ‘metaorganism’ (Bosch and McFall-Ngai 2011)) has been defined to cover the host plus its associated microbiota (Bordenstein and Theis 2015; Rohwer et al. 2002). While the histology and trophic ecology of glass sponges has been a matter of several previous studies (Kahn and Leys 2017; Kahn et al. 2018), the microbiology of glass sponges still remains largely unexplored (but see Steinert et al. 2020; Tian et al. 2016; Savoca et al. 2019).
The glass sponge Vazella pourtalesii (Schmidt 1870) is distributed along the continental margin of eastern North America from the Florida Keys in the southeastern USA to the Scotian Shelf off Nova Scotia, Canada, where it forms pronounced monospecific aggregations with densities reaching up to 4 individuals per m2 (Fig. 1; Fig. 2a, Kenchington et al. unpublished data). In 2013, Fisheries and Oceans Canada (DFO) established two Sponge Conservation Areas (the Sambro Bank and Emerald Basin Sponge Conservation Areas, referred to herein as SCAs) to protect two of the most significant concentrations of V. pourtalesii from bottom-fishing activities. However, these closures protect less than 2% of the total area covered by the V. pourtalesii sponge grounds (Kenchington et al. unpublished data), and bottom fishing activities continue to occur almost immediately adjacent to their borders (DFO 2017).
Map of sampling region on the Scotian Shelf, Canada. Colours depict the probability of V. pourtalesii occurrence based on the data presented in Beazley et al. (2018). Yellow indicates areas where the probability of occurrence is highest. The Sambro Bank and Emerald Basin SCAs are depicted by stripes. Sampling locations of V. pourtalesii in the Emerald Basin SCA are indicated by white dots and the white squares at the Sambro Bank SCA. The white triangle depicts the position of the OTN mooring and the white cross represents samples from the fisheries-impacted site
a In situ photographs of V. pourtalesii with individuals being up to around 20 cm in height. b Close-up of small V. pourtalesii growing on a mooring float of the Ocean Tracking Network (photo credits: DFO). Average size of small V. pourtalesii growing on the mooring was approximately 1 cm. Scale bar at right bottom represents 10 cm and applies only to the whole mooring float in Fig. 2b
Areas situated in the direct vicinity of fisheries closures represent potentially lucrative areas for fishing, as ‘spillover’ of fish stocks from the protected sites may occur. Bottom trawling is one of the most destructive ways to catch fish (Kelleher 2005). Impacts include reduced fishing stocks, by-catch of non-target species and destruction of habitat for benthic invertebrates in the trawl path and neighboring vicinity. At trawled sites, the local hydrodynamics may be altered due to a removal of habitat-forming benthic structures (living and non-living). Further, impacts at the population-, community,- and ecosystem-levels have resulted from the removals of unintentionally (bycaught) fished taxa (Ortuño Crespo and Dunn 2017). Benthic trawling also impacts the microbial community: For example Jackson et al. (2001) (and references therein) report a strong increase in microbial cell numbers in the water column associated with overfishing. For glass sponges such as V. pourtalesii, the effects of bottom trawling on the variability of microbial community compositions are currently unknown. In terms of sponge physiology, glass sponges were previously observed to arrest pumping in response to high sediment levels in the water column (Tompkins-Macdonald and Leys 2008), which may have an impact on the oxygenation status inside the sponge tissue and thus on the microbial community.
Several recent studies have explored the response of sponges and their microbiomes (hereafter referred to as ‘holobionts’) to anthropogenic stressors, such as ocean warming, acidification, eutrophication, sedimentation and pollution (reviewed in Pita et al. (2018); Slaby et al. (2019)). The increase in artificial substrates such as plastics in the oceans represents another anthropogenic-induced impact that could potentially adversely affect sponge holobionts. Especially in shelf areas, petroleum platforms, off-shore wind parks and increased loads of litter may represent new settlement opportunities for sponge larvae or gemmules. Artificial substrates have been observed to be exposed to microfouling and biofilm development (Balqadi et al. 2018). It is thus conceivable that biofilms on artificial substrates may influence the sponge holobiont settlement and development on such substrates. The recent discovery that V. pourtalesii uses oceanographic moorings (belonging to the Halifax Line of the Ocean Tracking Network (OTN)) as settlement substrate enabled the testing of this hypothesis. The present study aims to describe and compare the microbial community composition of the glass sponge V. pourtalesii collected from protected environments (i.e., within the Sambro Bank and Emerald Basin SCAs), a fisheries-impacted site immediately adjacent to the border of the Sambro Bank SCA, as well as from an artificial substrate (plastic mooring float cover). This study contributes to a deeper understanding of how marine sponge holobionts respond to human pressures.
Methods
Field work
Samples were collected during two oceanographic research missions to the Vazella sponge grounds on the Scotian Shelf, off Eastern Canada. The first mission took place in July/August 2016 onboard Canadian Coast Guard Ship (CCGS) Hudson (Hudson2016-019; Kenchington et al. 2017), where Oceaneering’s Spectrum remotely operated vehicle (ROV) and a mega-box corer were used to collect V. pourtalesii from the Emerald Basin SCA (protected site) between 184 and 206 m depth. The second mission was conducted in September 2017 onboard CCGS Martha L. Black (MLB2017001; Beazley et al. 2017), where sponge samples were collected during multiple deployments of the ROV ROPOS (Canadian Scientific Submersible Facility, Victoria, Canada) between 153 and 161 m depth at a single station near the centre of the Sambro Bank SCA (protected site). Furthermore, during this mission, sponge samples were collected outside the closure immediately adjacent to its southeast border at 198 m depth, in an area that was subjected to relatively recent bottom trawling activity (indicated by the presence of deep trawl marks in the general vicinity of the collection site). In total 33 sponges were collected: eight and 13 individuals from the Emerald Basin and Sambro Bank SCAs (protected sites), and four from the fisheries-impacted site adjacent to the Sambro Bank SCA. Additionally, eight specimens were collected from an Ocean Tracking Network (OTN) mooring located approximately 10 km northwest of the Sambro Bank SCA (for map of sampling locations see Fig. 1). The mooring (for a picture of the OTN mooring see Fig. 2b) was anchored ~ 5 m above the seabed and was deployed for ~ 13 months (15th of August 2016–5th of September 2017) prior to its recovery. Sample metadata were deposited in the Pangaea database.
From each V. pourtalesii specimen, pieces of sponge biomass were subsampled and rinsed by transferring them three times through sterile filtered seawater. Excess liquid was then removed, the samples were flash frozen, kept continuously frozen during transportation on dry-ice, and stored at − 80 °C until DNA extraction. For the mooring samples, complete sponge individuals were frozen at − 80 °C due to their small size.
Seawater samples (2 L) were collected (concomitantly with sponge sampling) at every location in quadruplicates and each filtered onto PVDF filter membranes (Merck Millipore) with a pore size of 0.22 µm and a diameter of 47 mm. Filters were stored at − 80 °C until DNA extraction. Sediments were collected using ROV push corers or box corer deployments at the same locations where sponges and seawater were collected contemporaneously. The top of each core was sliced off, the upper 2 cm of the sediment was collected and stored at − 80 °C until DNA extraction. Biofilms were scraped off the mooring float (where no visible encrusting fauna was present) using cotton swabs (Carl Roth, Karlsruhe, Germany), placed into sterile tubes and frozen at − 80 °C. All sponge, seawater, sediment and biofilm samples covered in this study originated from the same water mass (discussed in Beazley et al. 2018). Throughout this study we use the term ‘environmental references’ for seawater and sediment microbial communities. For assessing the impact of fisheries and artificial substrate provision on V. pourtalesii microbial communities, we use Sambro Bank protected sponges as the baseline for comparison.
Amplicon sequencing
DNA extraction was conducted on ~ 0.25 g of sponge tissue, ~ 0.25 g of sediment, half a seawater filter or the complete cotton woolen part of a swab using the DNeasy Power Soil Kit (Qiagen, Venlo, The Netherlands). Extracted DNA was quantified using Qubit fluorometer measurements and their quality assessed by a polymerase chain reaction (PCR) with the universal 16S rRNA gene primers 27F + 1492R and subsequent gel electrophoresis on 1% agarose. The V3 and V4 variable regions of the 16S rRNA gene were amplified in a one-step PCR using the primer pair 341F-806R in a dual-barcoding approach (Kozich et al. 2013). The nucleotide sequences of the primers are as follows: 5′-CCTACGGGAGGCAGCAG-3′ (Muyzer et al. 1993) and 5′-GGACTACHVGGGTWTCTAAT-3′ (Caporaso et al. 2011). PCR-products were verified by gel electrophoresis, normalised and pooled. Sequencing was performed on a MiSeq platform (MiSeqFGx, Illumina, San Diego, USA) using v3 chemistry (producing 2 × 300 bp). Demultiplexing after sequencing was based on 0 mismatches in the barcode sequences. Raw reads were archived in the NCBI Sequence Read Archive.
Bioinformatics and statistics
For analysis of sequencing data, raw sequences were first quality-filtered using BBDUK (BBMAP version 37.75; Bushnell 2017). Quality trimming was conducted on both ends of the reads (removal of first and last 13 nt) with Q20 and a minimum length of 250 nt. Quality of sequences was evaluated with FastQC (version 0.10.1; Andrews 2010) and output aggregated with MultiQC (version 0.9; Ewels et al. 2016). The post-filtered sequences were processed with QIIME2 (versions 2018.6 and 2018.8; Bolyen et al. 2019). The DADA2 algorithm (Callahan et al. 2016) was used, which retains Amplicon Sequence Variants (ASVs). Reads were denoised and consensus removal of chimera was conducted. One million reads were used to train the error model. Chloroplast and mitochondrial sequences were removed from further analyses. Assignment of taxonomy was conducted using a Naive Bayes classifier (Bokulich et al. 2018) trained on the Silva 132 99% OTUs 16S database (Quast et al. 2013). We used the term ‘unclassified’ which is defined as ‘no hit’ in this taxonomic classification. This information was used to identify potentially novel microbial diversity in V. pourtalesii. Ambiguous classifications (such as ‘unidentified bacteria’) are included in the ‘known sequences’ fraction as those sequence have been recorded and deposited before.
FastTree2 (Price et al. 2010) was used to generate a phylogeny based on ASVs. Subsequently, weighted UniFrac distances (phylogeny-based β-diversity metrices) were calculated (Lozupone et al. 2005). Non-metric multidimensional scaling (NMDS) was performed on weighted UniFrac distances to visually assess sample separation in ordination space. A permutational multivariate analysis of variance (PERMANOVA) was conducted on weighted UniFrac distances to determine whether groups of samples were significantly different from one another. Pairwise tests were performed between all pairs of groups (applied significant level α = 0.05). Multivariate analyses were run inside QIIME2 and R.
In addition to phylogeny-based metrics, quantitative measures of community richness were also calculated (e.g. Shannon’s Diversity Index). The Linear Discriminant Analysis (LDA) Effect Size (LEfSe) algorithm (Segata et al. 2011) was applied to determine if microbial phyla differed significantly among groups and rank them according to estimated effect sizes of the significant variations. To accomplish this, first a factorial Kruskal–Wallis sum-rank test was performed on feature tables to detect features (i.e. microbial taxa) with significant differential abundance between groups (applied significant level α = 0.05). Wilcoxon rank-sum tests were then run to perform pairwise tests among subgroups (applied significant level α = 0.05). Finally, a LDA estimated the effect size of each differentially abundant feature (applied threshold on the logarithmic LDA score for discriminative features = 2.0).
The majority of plots were produced using R statistical software version 3.0.2 (R Development Core Team 2008) and arranged using Inkscape (version 0.92.3; Harrington and Team 2005). The V. pourtalesii presence probability raster (Fig. 1) was visualised with QGIS (version 2.18.4; QGIS Development Team 2017).
Results
Microbial diversity of V. pourtalesii
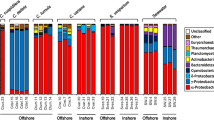
In the present study, we assessed the microbial community structure of 33 V. pourtalesii individuals as well as environmental reference samples by 16S rRNA gene amplicon sequencing. The microbiomes of the V. pourtalesii sponges clustered together, and were distinct from seawater and sediment microbiomes (Fig. 3). Overall, the V. pourtalesii microbiomes showed a higher variability of their α-diversity than the environmental reference microbiomes (seawater, biofilm, seawater). Rarefaction curves (α-diversity) revealed further that microbial richness (average number of ASVs per sample) was highest in the biofilm samples collected from the OTN mooring, second highest in seawater, and lowest in V. pourtalesii samples (Online Resource 1: S1). Also, in terms of diversity indices (Shannon index), alpha diversity was lowest in the V. pourtalesii microbiomes. While Proteobacteria were the most dominant phylum in all sample types (sponge: 78.8% of total; seawater: 47.4%; sediment: 54.7%; and biofilm: 56.6%), Patescibacteria were particularly abundant members of the V. pourtalesii microbiome (4.6%), followed by Bacteroidetes (3.4%), Spirochaetes (2.9%), and Planctomycetes (2.8%), (Fig. 4). Despite a distinct microbial fingerprint on different taxonomic levels (e.g., at ASV-level, as well as phylum-level), V. pourtalesii microbiomes showed a higher variability in terms of their α-diversity than environmental microbiomes. We further observed a higher proportion of previously unrecorded microbial taxa at every hierarchical taxonomic level (microbial phylum down to microbial order level) in the microbiomes of V. pourtalesii than in the seawater samples (Table 1). At the genus level, 40.2% of the amplicon sequences of V. pourtalesii could not be taxonomically assigned (compared to 16.2% in seawater). The largest increase of unclassified reads was observed from class to order level (from 0.4% to 31.6% respectively). In comparison to environmental microbiomes (seawater, sediment, and biofilm) V. pourtalesii microbiomes were particularly enriched in Proteobacteria and Patescibacteria (Fig. 5a).
Non-metric Multidimensional Scaling (NMDS) plot on weighted UniFrac distances. Each marker represents a single microbial community. Symbols represent sampling location: squares show samples from Sambro Bank, circles represent samples from Emerald Basin, triangles are mooring samples and crosses are samples from the fisheries impacted site. Colors depict sample type: green marks V. pourtalesii samples, blue marks seawater samples, yellow is for mooring biofilm and red represents sediment samples
Linear discriminant analysis (LDA) Effect Size (LEfSe) plots. Analysis was performed on microbial phylum level, the rank in the plot is given according to effect size. a Shows data for different sample types (sponge, seawater, sediment and mooring biofilm samples). Here, only the four most significantly different phyla are shown per group. For b-d, where only sponge samples were inspected, all phyla are shown. b Shows differences between sponge conservation areas (SCAs), Sambro Bank and Emerald Basin. c-d) Show microbial taxa increased or decreased upon anthropogenic pressures. c Shows protected vs. fisheries-impacted sites and d shows growth of sponges in their natural habitat in sponge grounds vs moorings). For all plots, filled bars show microbial taxa with significant effects only in sponge samples; striped bars additionally indicate that these taxa have a significant effect for all three sample types (sponge, seawater and sediment). Dots next to or below plots indicate relative abundance of taxa within the V. pourtalesii microbial community. The respective scale is indicated at the lower part of the illustration
Response of the V. pourtalesii microbiome to anthropogenic activities
We then assessed the changes of the sponge microbiome upon human activity, including also implemented protection efforts in form of sponge conservation areas. A significant difference in V. pourtalesii microbiomes between the Sambro Bank and Emerald Basin SCAs was observed (Table 2), with several microbial phyla significantly enriched in one site over the other (Fig. 5b). For example, Patescibacteria were significantly enriched in Sambro Bank sponges, while Proteobacteria were significantly enriched in Emerald Basin sponges. We further compared inter-location variability (i.e., V. pourtalesii individuals between the Emerald Basin SCA and Sambro Bank SCA) against intra-location variability (i.e., individuals collected from within Emerald Basin SCA only). While the inter-location variability was significant, the intra-location variability was not (PERMANOVA, Online Resource 1: Table S1, mean weighted UniFrac distance = 0.29). Further, a smaller mean weighted UniFrac distance was observed between sponges of the same location (Emerald Basin SCA), compared to sponges at different sampling locations (Emerald Basin SCA vs. Sambro Bank SCA). In terms of trawling impact, certain microbial taxa were depleted or enriched in V. pourtalesii individuals originating from the trawled site compared to the protected sites (Fig. 5c). For example, Nitrospinae were significantly depleted in V. pourtalesii individuals originating from the trawled site adjacent to the Sambro Bank SCA (in both sponges as well as environmental references). Nitrospinae are nitrite-oxidising bacteria and have previously been found in deep-sea glass sponges (Tian et al. 2016). With respect to sponge growth on artificial substrates (moorings), their microbial community composition differed significantly from sponges growing in their natural habitat (Table 2). Sponges from the artificial mooring substrate were significantly enriched in Bacteroidetes, Verrucomicrobia and Cyanobacteria which were also enriched in the mooring biofilm (Fig. 5d).
Discussion
This study is the first to report on the microbiome composition of the glass sponge Vazella pourtalesii, that forms pronounced monospecific aggregations with high densities on the Scotian Shelf, Canada (Fuller 2011; Beazley et al. 2018) that are not found elsewhere in the world’s oceans. Our results suggest that V. pourtalesii contains its own distinct microbial community that is different from seawater and sediment samples. The observed pattern of lower alpha diversity in V. pourtalesii compared to environmental reference samples is in line with previous observations showing that sponge-associated microbial communities are usually less complex than those of seawater or sediments (Thomas et al. 2016). We propose that V. pourtalesii represents a rich reservoir of novel microbial taxa, even at high taxonomic ranks (e.g. class-level: 31.6% unclassified). The high abundance of Patescibacteria in V. pourtalesii is particularly striking when compared to the microbiomes of other glass sponges (Tian et al. 2016) and also when compared to the environmental reference samples in our study. This microbial phylum is generally assumed to depend on symbiotic animal hosts to cover their basic metabolic requirements (Castelle et al. 2018).
V. pourtalesii microbial community composition varies between the Sambro Bank and Emerald Basin Sponge Conservation Areas
Compared to the conservation efforts towards the protection of animal and plant biodiversity, comparably little is currently known about the needs to protect microbial biodiversity. This is striking as Webster et al. (2018) and others point out very clearly that microorganisms underpin ecosystem health and hence should deserve committed conservation and management endeavors. In our study, we explored the variation in microbial community composition residing in sponges from two SCAs. We observed that the variations in sponge-associated microbiomes were smaller within sites than between sites. We cannot rule out whether a temporal effect may have led to the observed differences between sites, as all Emerald Basin samples were collected in 2016, while all other samples were collected in 2017. However, since no significant differences in seawater microbial communities between both years were observed (Table 2), we deduce that temporal effects were not detectable in the water surrounding the sponges. Our results of variable microbial community compositions between Emerald Basin SCA V. pourtalesiis and Sambro Bank SCA V. pourtalesiis illustrate that fisheries closures are important for the conservation of not only macro-, but also microbial biodiversity.
Deviations of the V. pourtalesii microbial community composition at a trawled site
We observed significantly different relative abundances of distinct microbial phyla from bottom-trawled areas, such as a reduction of Nitrospinae in sponges and reference samples. On the other hand, Planctomycetes, Actinobacteria as well as Tenericutes were significantly enriched in V. pourtalesii individuals originating from the fisheries-impacted site, but not in the seawater or other samples taken from these sites. Although we observed significant enrichments or depletions of individual microbial phyla, the overall V. pourtalesii microbial community composition did not differ significantly between the protected and fisheries-impacted site in a permanova analyses based on weighted UniFrac distances. As outcomes of permanova analyses strongly depend on sample sizes, we cannot exclude a technical issue in this regard (as n = 4). However, these results are in line with Luter et al. (2012) who observed no shifts in the microbial community of the Great Barrier reef sponge Ianthella basta associated with increased sedimentation, when considering the total microbial community rather than individual microbial taxa. In another study on five shallow water sponge species, again no significant effect of increased sediment loads was observed, while minor effects included an increased reliance on phototrophic feeding under high suspended sediment loads (Pineda et al. 2016). Future studies with targeted sampling, higher replication, and wet-lab experimentation are needed to fully assess the impact of trawling on deep-sea sponges and their microbiomes.
While seawater and sediment microbiomes differed significantly from each other at the protected sites in our study, microbial community composition was similar for the two sample types at the fisheries-impacted site. Seawater microbial community composition was significantly different at the fisheries-impacted site in comparison to the protected site, while the sediment microbial community was similar at both sites. From these two observations we conclude that trawling might have an effect on the seawater microbial community in a way that the seawater community becomes more similar to that of sediments. We posit that this may be due to direct and indirect effects of trawling-induced sediment suspension. While a direct translocation of microbial community members may occur from the sediment to the seawater, sediment resuspension may also lead to an increased eutrophication, thus promoting a rise of opportunistic bacterial clades in response to the release of biogeochemical elements (such as e.g., phosphorous) from the sediment into the water column.
Deviations of the V. pourtalesii microbial community composition growing on an artificial substrate
Artificial structures have the potential to act as stepping stones, enabling species to broaden their distributions in the marine environment (Adams et al. 2014). Our results suggest that the biofilms on artificial substrates may ‘prime’ the sponge-associated microbial communities when small sponges settle on such substrates. Our finding of a different microbial community in mooring V. pourtalesii individuals compared to protected ones raises the possibility of these structures changing the microbial diversity of adjacent and newly established benthic populations seeded from such sources. Individuals of V. pourtalesii growing on the artificial substrate showed a higher degree of microbiome variation compared to those collected from their natural habitat (Online Resource 1: S2). This variation may be due to the fact that sponges growing on the moorings were still small and had presumably settled within a maximum of 13 months following mooring deployment, and thus their microbiomes may not yet have been fully established.
Conclusions
In the present study two anthropogenic impacts, exposure to trawling activity and growth on plastic moorings, on the V. pourtalesii microbiomes were explored. In addition, we evaluated the role of two fisheries closures in preserving sponge-associated microbial diversity on the Scotian Shelf, Canada. We conclude first and foremost, that the Emerald Basin and Sambro Bank Sponge Conservation Areas are ecologically important, as they contain distinct and largely unclassified benthic microbial communities found within sponges or sediments. Variability of the microbial community was comparably high between V. pourtalesii individuals, pointing towards the need to minimise sponge loss through anthropogenic pressures (e.g., direct trawling) in order to protect the full range of microbial biodiversity. We further observed significant quantitative differences (in terms of relative abundances) in distinct microbial phyla, such as a reduction in Nitrospinae in sponges and samples from the surrounding environment in areas subjected to bottom trawling. With respect to sponges growing on plastic moorings, significant differences in their microbiome were observed compared to sponges growing on natural substrates. V. pourtalesii individuals growing on a mooring surface were significantly enriched in Bacteroidetes, Verrucomicrobia and Cyanobacteria compared to those growing on natural substrates. This could be explained by either the plastic settlement substrate or the younger age of sponges growing on the moorings. Importantly, V. pourtalesii is a rich and unique reservoir of microbial biodiversity that deserves conservation effort.
Data availability
Sample metadata were deposited in the Pangaea database: https://doi.pangaea.de/10.1594/PANGAEA.913907. Raw sequences were archived in the NCBI Sequence Read Archive under BioProject id: PRJNA613976.
References
Adams TP, Miller RG, Aleynik D, Burrows MT (2014) Offshore marine renewable energy devices as stepping stones across biogeographical boundaries. J Appl Ecol 51(2):330–338
Andrews S (2010) FastQC: a quality control tool for high throughput sequence data. Available online at: https://www.bioinformatics.babraham.ac.uk/projects/fastqc.
Balqadi AA, Salama AJ, Satheesh S (2018) Microfouling development on artificial substrates deployed in the central Red Sea. Oceanologia 60:219–231
Beazley L, Pham C, Murillo J, Kenchington E (2017) Cruise report for the DFO/SponGES CCGS Martha L. Black oceanographic mission (MLB2017001), August 31 to September 7, 2017. Canadian Technical Report of Fisheries and Aquatic Sciences 3242: vi+42.
Beazley L, Wang Z, Kenchington E, Yashayaev I, Rapp HT, Xavier JR et al (2018) Predicted distribution of the glass sponge Vazella pourtalesi on the Scotian Shelf and its persistence in the face of climatic variability. PLoS One 13:e0205505
Bokulich NA, Kaehler BD, Rideout JR, Dillon M, Bolyen E, Knight R et al (2018) Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6:1–17
Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA et al (2019) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857. https://doi.org/10.1038/s41587-019-0209-9
Bordenstein SR, Theis KR (2015) Host biology in light of the microbiome: ten principles of holobionts and hologenomes. PLoS Biol 13:e1002226
Bosch TCG, McFall-Ngai MJ (2011) Metaorganisms as the new frontier. Zoology 114:185–190
Bushnell B (2017) BBMap short read aligner, and other bioinformatic tools. Available online at: https://sourceforge.net/projects/bbmap/.
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP (2016) DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Knight R, Lozupone CA et al (2011) Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci 108:4516–4522
Castelle CJ, Brown CT, Anantharaman K, Probst AJ, Huang RH, Banfield JF (2018) Biosynthetic capacity, metabolic variety and unusual biology in the CPR and DPANN radiations. Nat Rev Microbiol 16:629–645
DFO (2017) Delineation of significant areas of coldwater corals and sponge-dominated communities in Canada’s Atlantic and Eastern Arctic marine waters and their overlap with fishing activity. DFO Can Sci Advis Sec Sci Advis Rep 2017:007
Ewels P, Magnusson M, Lundin S, Käller M (2016) MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32:3047–3048
Fuller S (2011) Diversity of marine sponges in the Northwest Atlantic. PhD Thesis, Dalhousie Univ 1–229.
Harrington B and Team, and the D. (2005) Inkscape. Available online at: https://www.inkscape.org/.
Jackson JBC, Kirby MX, Berger WH, Bjorndal KA, Botsford LW, Bourque BJ et al (2001) Historical overfishing and the recent collapse of coastal ecosystems. Science (80–) 629:1–17
Kahn AS, Chu JWF, Leys SP (2018) Trophic ecology of glass sponge reefs in the Strait of Georgia, British Columbia. Sci Rep 8:1–11
Kahn AS, Leys SP (2017) Spicule and flagellated chamber formation in a growth zone of Aphrocallistes vastus (Porifera, Hexactinellida). Invertebr Biol 136:22–30
Kelleher K (2005) Discards in the world’s marine fisheries. An update. FAO Fisheries Technical Paper. 470.
Kenchington E, Beazley L, Yashayaev I (2017) Hudson 2016–019 International deep sea science expedition cruise report. Canadian Data Report of Fisheries and Aquatic Sciences 1277: v + 55p.
Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD (2013) Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the miseq illumina sequencing platform. Appl Environ Microbiol 79:5112–5120
Leys SP, Mackie GO, Reiswig HM (2007) The biology of glass sponges. Adv Mar Biol 52:1–145
Lozupone C, Knight R, Lozupone C, Knight R (2005) UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71:8228–8235
Luter HM, Whalan S, Webster NS (2012) Thermal and sedimentation stress are unlikely causes of brown spot syndrome in the coral reef sponge, Ianthella basta. PLoS One 7:1–9
McFall-Ngai M, Hadfield MG, Bosch TCG, Carey HV, Domazet-Lošo T, Douglas AE et al (2013) Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci USA 110:3229–3236
Moitinho-Silva L, Nielsen S, Amir A, Gonzalez A, Ackermann GL, Cerrano C, et al (2017) The sponge microbiome project. Gigascience 6(10):gix077
Muyzer G, Waal ECDE, Uitierlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:1–6
Ortuño Crespo G, Dunn DC (2017) A review of the impacts of fisheries on open-ocean ecosystems. ICES J Mar Sci 74:2283–2297
Pineda M-C, Strehlow B, Duckworth A, Doyle J, Jones R, Webster NS (2016) Effects of light attenuation on the sponge holobiont- implications for dredging management. Sci Rep 6:39038
Pita L, Rix L, Slaby BM, Franke A, Hentschel U (2018) The sponge holobiont in a changing ocean: from microbes to ecosystems. Microbiome 6:46
Price MN, Dehal PS, Arkin AP (2010) FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490
QGIS Development Team (2017) Geographic information system. Open source geospatial foundation project. Available online at: https://qgis.osgeo.org.
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P et al (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:590–596
R Development Core Team (2008) R: a language and environment for statistical computing.
Rohwer F, Seguritan V, Azam F, Knowlton N (2002) Diversity and distribution of coral-associated bacteria. Mar Ecol Prog Ser 243:1–10
Savoca S, Lo Giudice A, Papale M, Mangano S, Caruso C, Spanò N et al (2019) Antarctic sponges from the Terra Nova Bay (Ross Sea) host a diversified bacterial community. Sci Rep 9:1–15
Schmidt O (1870) Grundzüge einer Spongien-Fauna des atlantischen Gebietes. Wilhelm Engelmann, Leipzig, pp 14–15
Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C (2011) Metagenomic biomarker discovery and explanation. Genome Biol 12:R60
Slaby B, Franke A, Rix L, Pita L, Bayer K, Jahn M, Hentschel U (2019) Marine sponge holobionts in health and disease. In: Li Z (ed) Symbiotic microbiomes of coral reefs sponges and corals. Springer, Netherlands, Dordrecht, pp 81–104
Steinert G, Busch K, Bayer K, Khodami S, Arbizu PM, Kelly M, et al (2020) Compositional and quantitative insights into bacterial and archaeal communities of South Pacific deep-sea sponges (Demospongiae and Hexactinellida). Front Microbiol 11:716
Thomas T, Moitinho-Silva L, Lurgi M, Björk JR, Easson C, Astudillo-García C et al (2016) Diversity, structure and convergent evolution of the global sponge microbiome. Nat Commun 7:11870
Tian RM, Sun J, Cai L, Zhang WP, Zhou GW, Qiu JW, Qian PY (2016) The deep-sea glass sponge Lophophysema eversa harbours potential symbionts responsible for the nutrient conversions of carbon, nitrogen and sulfur. Environ Microbiol 18:2481–2494
Tompkins-Macdonald GJ, Leys SP (2008) Glass sponges arrest pumping in response to sediment: implications for the physiology of the hexactinellid conduction system. Mar Biol 154:973–984
Webster NS, Wagner M, Negri AP (2018) Microbial conservation in the Anthropocene. Environ Microbiol 20:1925–1928
Acknowledgements
Open Access funding provided by Projekt DEAL. This manuscript is written in memory of Hans Tore Rapp, whose research efforts were dedicated to the understanding and preservation of deep-sea sponge grounds in the North Atlantic Ocean. We thank the crews and scientific parties of the two research cruises Hudson2016-019 and MLB2017001. Ulrike Hanz and Furu Mienis are acknowledged for support during field work. We further thank Andrea Hethke, Ina Clefsen, and the CRC1182 Z3 team (Katja Cloppenborg-Schmidt, Malte Rühlemann, John Baines) for assistance with the amplicon pipeline.
Funding
This study was funded by the European Union’s Horizon 2020 research and innovation program under Grant Agreement No. 679849 (‘SponGES’). This document reflects only the authors’ view and the Executive Agency for Small and Medium-sized Enterprises (EASME) is not responsible for any use that may be made of the information it contains. Canadian cruises and contributions were funded by Fisheries and Ocean’s Canada’s International Governance Strategy Science Program through project “Marine Biological Diversity Beyond Areas of National Jurisdiction (BBNJ): 3-Tiers of Diversity (Genes-Species-Communities)” led by EK (2017–2019).
Author information
Authors and Affiliations
Contributions
KB, UH and BS designed the study. EK, LB, FW organised the cruises and provided samples. KB performed material preparations and data analyses (bioinformatics and visualisation). The first draft of the manuscript was written by KB and UH. All authors commented on previous versions of the manuscript and approved the final version.
Corresponding author
Ethics declarations
Conflict of interests
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Busch, K., Beazley, L., Kenchington, E. et al. Microbial diversity of the glass sponge Vazella pourtalesii in response to anthropogenic activities. Conserv Genet 21, 1001–1010 (2020). https://doi.org/10.1007/s10592-020-01305-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-020-01305-2