Abstract
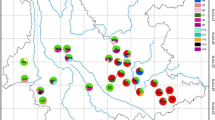
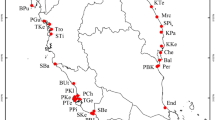
Rosa praelucens is a critically endangered decaploid alpine rose with an extremely narrow geographic distribution in Northwestern Yunnan, China. We sampled almost all the extant individuals (527 individuals in 31 natural locations and 56 individuals preserved in three local living collections) to assess the genetic variation and to probe the genetic connectivity among the individuals and populations based on three cpDNA intergenic spacers and six fluorescent amplified fragment length polymorphism (AFLP) markers. The morphological traits from seven populations were also measured. R. praelucens exhibited high levels of morphological variation, genetic diversity, and differentiation. The extant individuals were clustered into eight groups in neighbor-net networks, and subsequent Bayesian analysis assigned them into three larger gene pools, both in accordance with their morphological traits, especially flower color. The living collections embraced two private cpDNA haplotypes and included three out of the species’ total eight AFLP genotypes. Rhizome clonal growth, decaploid, and mixed breeding system may largely contribute to high genetic diversity and differentiation in R. praelucens. We concluded that the endangered status of R. praelucens may mainly be due to habitat fragmentation and loss and inherent reproductive difficulties, rather than low genetic diversity. The populations contributing higher cpDNA genetic diversity, representing more AFLP genotypes, and encompassing private cpDNA haplotypes should be given conservation priority by creating plant-micro reserves. The living collections should also be targeted for further ex situ conservation, population recovery, and reintroduction of R. praelucens plants.





Similar content being viewed by others
References
Byhouwer JTP (1929) An enumeration of the roses of Yunnan. J Arnold Arbor 10:84–107
De Cock K, Vander Mijnsbrugge K, Breyne P, Van Bockstaele E, Van Slycken J (2008) Morphological and AFLP-based differentiation within the taxonomical complex Section Caninae (subgenus Rosa). Ann Bot 102:685–697
Demesure B, Sodzin, Petit RJ (1995) A set of universal primers for amplification of polymorphic non-coding regions of mitochondrial and chloroplast DNA in plants. Mol Ecol 4:129–131
Deng JQ, Jian HY, Li SB, Wang QG, Guo YL, Zhang H (2013) Cold tolerance of several wild rose resources endemic to Yunnan. Southwest China J Agric Sci 26:723–727
Despres L, Loriot S, Gaudeul M (2002) Geographic pattern of genetic variation in the European globeflower Trollius europaeus L. (Ranunculaceae) inferred from amplified fragment length polymorphism markers. Mol Ecol 11:2337–2347
Dufresne F, Stift M, Vergilino R, Mable B (2014) Recent progress and challenges in population genetics of polyploidy organisms: an overview of current state-of-the-art molecular and statistical tools. Mol Ecol 23:40–69
Dupanloup I, Schneider S, Excoffier L (2002) A simulated annealing approach to define the genetic structure of populations. Mol Ecol 11:2571–2581
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinform Online 1:47–50
Fougère-Daneza M, Joly S, Bruneau A, Gao XF, Zhang LB (2014) Phylogeny and biogeography of wild roses with specific attention to polyploids. Ann Bot 115:275–291
Fu YX (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147:915–925
Furches MS, Small RL, Rurches A (2013) Genetic diversity in three endangered pitcher plant species (Sarracenia; Sarraceniaceae) is lower than widespread congeners. Am J Bot 100:2092–2101
Gaudeul MP, Taberlet MP, Till-Bottraud I (2000) Genetic diversity in an endangered alpine plant, Eryngium alpinum L. (Apiaceae), inferred from amplified fragment length polymorphism markers. Mol Ecol 9:1625–1637
Gitzendanner MA, Soltis PS (2000) Patterns of genetic variation in rare and widespread plant congeners. Am J Bot 87:783–792
Goossens B, Sharma R, Othman N et al (2016) Habitat fragmentation and genetic diversity in natural populations of the Bornean elephant: implications for conservation. Biol Conserv 196:80–92
Graur D, Li WH (2000) Fundamentals of molecular evolution, 2nd edn. Sinauer Associates, Sunderland
Guan WL, Li SF, Song J, Pan LJ, Niu HB (2012) Study on geographic distribution of Rosa praelucens endemic to Yunnan. J West China For Sci 41:88–93
Gutin S (2003) Seed propagation. In: Robersts AV, Debener T, Gudin S (eds) Encylopedia of rose science. Academic Press, Elsevier, London, pp 620–623
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hamilton MB (1999) Four primer pairs for the amplification of chloroplastic intragenic regions with intraspecific variation. Mol Ecol 8:513–525
Hamrick JL, Godt MJW (1989) Allozyme diversity in plant species. In: Brown AHD, Clegg MT, Kahler AL, Weir BS (eds) Plant population genetics, breeding, and genetic resources. Sinauer Associates, Sunderland, pp 43–63
Hamrick JL, Godt MJW (1996) Effects of life history traits on genetic diversity in plant species. Philos Trans R Soc Lond B 351:1291–1298
Hardy OJ, Vekemans X (2001) Patterns of allozyme variation in diploid and tetraploid Centaurea jacea at different spatial scales. Evolution 55:943–954
Honnay O, Bossuyt B (2005) Prolonged clonal growth: escape route or route to extinction? Oikos 108:427–432
Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755
Huson DH, Bryant D (2006) Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23:254–267
Ikeda H, Setoguchi H (2009) The homogenous genetic structure and inferred unique history of range shifts during the Pleistocene climatic oscillations of Arcterica nana (Maxim.) Makino (Ericaceae). J Plant Res 122:141–151
Jian HY, Zhang H, Tang KX et al (2010a) Decaploidy in Rosa praelucens Byhouwer (Rosaceae) endemic to Zhongdian Plateau, Yunnan, China. Caryologia 63:162–167
Jian HY, Zhang H, Wang QG et al (2010b) Karyological study of Chinese old garden roses. Acta Hortic Sin 37:83–88
Jian HY, Tang KX, Sun H (2015) Phylogeography of Rosa soulieana (Rosaceae) in the Hengduan Mountains: refugia and ‘melting’ pots in the Quaternary climate oscillations. Plant Syst Evol 301:1819–1830
Jian HY, Zhang YH, Qiu XQ et al (2016) Yalongjiang river has had an important role in the dispersal and divergence of Rosa soulieana in the Hengduan Mountains of China. PLoS ONE 11:e0158586. https://doi.org/10.1371/journal.pone.0158586
Jian HY, Zhang SD, Zhang T et al (2017) Characterization of the complete chloroplast genome of a critically endangered decaploid rose species, Rosa praelucens (Rosaceae). Conserv Genet Resour. https://doi.org/10.1007/s12686-017-0946-3
Karron JD (1987) A comparison of levels of genetic polymorphism and self-compatibility in geographically restricted and widespread plant congeners. Evol Ecol 1:47–58
Kato S, Iwata H, Tsumura Y, Mukai Y (2011) Genetic structure of island populations of Prunus lannesiana var. speciosa revealed by chloroplast DNA, AFLP and nuclear SSR loci analyses. J Plant Res 124:11–23
Kellner A, Ritz CM, Vissemann V (2014) Low genetic and morphological differentiation in the European species complex of Rosa sherardii, R. mollis and R. villosa (Rosa section Caninae subsection Vestitae). Bot J Linn Soc 174:240–256
Ku TC, Robertson KR (2003) Rosa (Rosaceae). In: Wu ZY, Raven PH (eds) Flora of China. Science Press, Beijing, pp 339–380
Laguna E (2001) The micro-reserves as a tool for conservation of threatened plants in Europe. Council of Europe, Strabourg
Laguna E, Deltoro VI, Perez-Botella J, Perez-Rovira P, Olivares A, Fabregat C (2004) The role of small reserves in plant conservation in a region of high diversity in eastern Spain. Biol Conserv 119:421–426
Li SF (2012) Key techniques in the domestication and cultivation of Rosa praelucens Byhouwer in Kunming. Dissertation, Graduate school of Chinese Academy of Agricultural Sciences
Li XX, Zhou ZK (2005) Endemic wild ornamental plants from Northwestern Yunnan., China HortScience 40:1612–1619
Li Y, Zhai SN, Qiu YX, Guo YP, Ge XJ, Comes HP (2011) Glacial survival east and west of the ‘Mekong–Salween-Divide’ in the Himalaya-Hengduan Mountains region as revealed by AFLP and cpDNA sequence variation in Sinopodophyllum hexandrum (Berberidaceae). Mol Phylogenet Evol 59:412–424
Li SF, Li CJ, Jian HY, Li SB et al (2013) Studies on phenotypic diversity of vulnerable Rosa praelucens endemic to Shangrila, Yunnan. Acta Hortic Sin 40:924–932
Liu DH, Li MX (1985) A study on karyotypes of some flowers of Rosa in China. J Wuhan Bot Res 3:403–408
Liu J, Sun P, Zheng XY et al (2013) Genetic structure and phylogeography of Pyrus pashia (Rosaceae) in Yunnan Province, China, revealed by chloroplast DNA analyses. Tree Genet Genom 9:433–441
Mason CM, Ishibashi CDA, Rea AM, Mandel JR, Burke JM, Donovan LA (2015) Environmental requirements trump genetic factors in explaining narrow endemism in two imperiled Florida sunflowers. Conserv Genet 16:1277–1293
Moody ME, Mueller LD, Soltis DE (1993) Genetic variation and random genetic drift in autotetraploid populations. Genetics 134:649–657
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Nei M, Tajima F (1983) Maximum likelihood estimation of the number of nucleotide substitutions from restriction sites data. Genetics 105:207–217
Nybom H (2004) Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol Ecol 13:1143–1155
Nybom H, Bartish IV (2000) Effect of life history traits and sampling strategies on genetic diversity estimates obtained with RAPD markers in plants. Perspect Plant Ecol Evol Syst 3:93–114
Pan LJ, Guan WL, Hua WH, Niu HB (2012) Community characteristics of endangered species Rosa praelucens. Subtrop Plant Sci 41:51–55
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for research and teaching. Mol Ecol Notes 6:288–295
Petit RJ, Hampe A (2006) Some evolutionary consequences of being a tree. Ann Rev Ecol Evol Syst 37:187–214
Petit R, Mousadik El A, Pons O (1998) Identifying populations for conservation on the basis of genetic markers. Conserv Biol 12:844–855
Petit RJ, Duminil J, Fineschi S, Hampe A, Salvini D, Vendramin GG (2005) Comparative organization of chloroplast, mitochondrial and nuclear diversity in plant populations. Mol Ecol 14:689–701
Pons O, Petit RJ (1996) Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics 144:1237–1245
Posada D, Crandall KA (1998) Modeltest: testing the model of DNA substitution. Bioinformatics 14:817–818
Prichard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Qin HN, Yang Y, Dong SY et al (2017) Threatened species list of China’s higher plants. Biodiv Sci 25:696–744
Ramsey J, Schemske DW (1998) Pathways, mechanisms, and rates of polyploidy formation in flowering plants. Annu Rev Ecol Syst 29:467–501
Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R (2003) DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496–2497
Segraves K, Thompson J, Soltis P, Soltis D (1999) Multiple origins of polyploidy and the geographic structure of Heuchera grossulariifolia. Mol Ecol 8:253–262
Shaw J, Lickey EB, Beck JT et al (2005) The tortoise and the hare II: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Am J Bot 92:142–166
Shaw J, Lickey EB, Schilling EE, Small RL (2007) Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in Angiosperms: the tortoise and the hare III. Am J Bot 94:275–288
Sobel JM, Streifeld MA (2013) Flower color as a model system for studies of plant evo-devo. Front Plant Sci 4:321. https://doi.org/10.3389/fpls.2013.00321
Soltis PS, Soltis DE (2009) The role of hybridization in plant speciation. Annu Rev Plant Biol 60:561–588
Stelling MA, van der Peijl GLQ (2001) Forensic methods for criminal investigations related to endangered species. Netherlands Forensic Institute, Rijswijk
Swofford DL (2000) PAUP4.0b10. Phylogenetic analysis using parsimony and other methods, vol 4. Sinauer Associates, Sunderland
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL X windows interface: flexible strategies for multiple sequences alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Ueda Y (2003) Seed maturation and germination. In: Robersts AV, Debener T, Gudin S (eds) Encyclopedia of rose science. Academic Press, Elsevier, London, pp 623–626
Vander Mijnsbrugge K, De Cock K, Cox K, Breyne P (2010) Conservation measures for Rosa arvensis Huds. in Flanders (Belgium) based on congruent genetic and phenotypic population differentiation. Conserv Genet 11:2243–2253
Volis S (2016) How to conserve threatened Chinese plant species with extremely small populations? Plant Diver 1:1–11
Vos P, Hogers R, Bleeker M et al (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Wang S, Xie Y (2004) China species red list, vol 1. Higher Education Press, Beijing
Webb T, Bartlein PJ (1992) Global changes during the last 3 million years: climatic controls and biotic response. Annu Rev Ecol Syst 23:141–173
Williamson PS, Werth CR (1999) Levels and patterns of genetic variation in the endangered species Abronia macrocarpa (Nyctaginaceae). Am J Bot 86:293–301
Wissemann V (2003) Conventional taxonomy (wild roses). In: Roberts AV, Debener T, Gudin S (eds) Encyclopedia of rose science. Elsevier, Amsterdam, pp 111–117
Wu XY, Chen M, Wang QG et al (2014) Comparative study on the breeding systems of Rosa praelucens and Rosa soulieana. Acta Hortic Sin 41:2075–2084
Yeh FC, Yang RC, Boyle TBJ, Ye ZH, Mao JX (1997) POPGENE, the user friendly shareware for population genetic analysis. Molecular Biology and Biotechnology Centre. University of Alberta, Edmonton
Young AB, Brown AHD (1996) Comparative population genetic structure of the rare woodland shrub Daviesia suaveolens and its common congener D mimosoides. Conserv Biol 10:1220–1228
Zhou YQ, Su Q, Zhang H, Li SF, Tang KX, Jian HY (2016) Distribution and population quantitative dynamics of critically risked Rosa praelucens Byhouwer. J Plant Genet Resour 17:649–654
Zozomová-Lihová J, Malánová-Krásná I, Vít P et al (2015) Cytotype distribution patterns, ecological differentiation, and genetic structure in a diploid-tetraploid contact zone of Cardamine amara. Am J Bot 102:1380–1395
Acknowledgements
Yuquan Zhou, Qun Su, Hao Zhang and Cankun Xiong helped to collect samples. This work was financially supported by the Grants from National Natural Science Foundation of China (31260198, 31560301) and the Academic and Technical Talent Training Project of Yunnan Province, China (2013HB092).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10592_2018_1052_MOESM3_ESM.eps
Values of differentiation among groups of population (Fct) obtained from SAMOVA as a function of the user-defined number of groups (k) (EPS 611 KB)
10592_2018_1052_MOESM4_ESM.eps
Distribution of the number of pairwise nucleotide differences for cpDNA haplotypes in R. praelucens. Dashed line indicates observed values (Obs). Solid line represents expected values (Exp) under a model of sudden population expansion (EPS 406 KB)
10592_2018_1052_MOESM6_ESM.eps
Genetic structure of R. praelucens based on the Bayesian analysis. (A) Log probability of data L(k) as function of k for 10 runs from k=2 to 20. (B) Rate of change in the probability between the successive runs, Δk, as a function of k, in the Bayesian analysis, indicating K=3. (C) Assignment of 583 individuals from 34 populations into 3 (k=3) genetically distinguished groups. Each individual is represented by a vertical bar partitioned into 3 colored segments that denotes the individuals’ estimated membership fraction in 3 clusters (EPS 2201 KB)
Rights and permissions
About this article
Cite this article
Jian, H., Li, S., Guo, J. et al. High genetic diversity and differentiation of an extremely narrowly distributed and critically endangered decaploid rose (Rosa praelucens): implications for its conservation. Conserv Genet 19, 761–776 (2018). https://doi.org/10.1007/s10592-018-1052-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-018-1052-0