Abstract
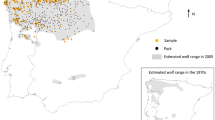
We studied the population structure and historical demography of the last remaining core population of the threatened southeastern beach mouse (SEBM; Peromyscus polionotus niveiventris) located on a federally protected barrier island complex at the Kennedy Space Center (KSC), Merritt Island National Wildlife Refuge (MINWR) and Cape Canaveral Air Force Station (CCAFS) in Florida, USA. Beach mice (N = 171) were collected from 33 trapping locations along 30 km of coastline on KSC/MINWR/CCAFS and were genotyped using 10 microsatellite loci. We found four genetic clusters of mice that likely form a metapopulation. Gene flow among clusters, assessed using assignment tests, was limited suggesting that human development can serve to inhibit dispersal of beach mice. However, when the presence of roads were examined as possible barriers to movement, gene flow appeared to be facilitated suggesting that removal of thick vegetation along roadsides increases movement. We used approximate Bayesian computation (ABC) to estimate divergence time among clusters and effective population sizes for each cluster and for the pre-divergence population. Results of ABC analyses suggest that barriers to movement likely formed following the construction of the John F. Kennedy Space Center beginning in the 1960s but that this has not heavily impacted the effective size of populations. Pre-divergence and contemporary effective sizes are similar, thus, population sizes likely remained relatively large over the last 50–100 years. The population of SEBM on KSC/MINWR/CCAFS appears to be a genetically diverse core population and individuals from this population will most likely be good candidates for any future reintroduction and translocation programs.



Similar content being viewed by others
References
Arnold TW (2010) Uninformative parameters and model selection using Akaike’s information criterion. J Wildl Manag 74:1175–1178
Bates D, Maechler M, Bolker B, Walker S (2014) lme4: linear mixed-effects models using Eigen and S4. R package ver 1.1–6. http://CRAN.R-project.org/package=lme4
Beaumont MA, Zhang W, Balding DJ (2002) Approximate Bayesian computation in population genetics. Genetics 162:2025–2035
Berry OM, Tocher D, Sarre SD (2004) Can assignment tests measure dispersal? Mol Ecol 13:551–561
Bertorelle G, Benazzo A, Mona S (2010) ABC as a flexible framework to estimate demography over space and time: some cons, many pros. Mol Ecol 19:2609–2625
Bissonette JA, Rosa SA (2009) Road zone effects in small-mammal communities. Ecol Soc 14:27
Bonnell M, Selander R (1974) Elephant seals: genetic variation and near extinction. Science 184:908–909
Bouzat J, Lewin H, Paige K (1998) The ghost of genetic diversity past: historical DNA analysis of the greater prairie chicken. Am Nat 152:1–6
Cain ML, Bowman WD, Hacker SD (2011) Ecology, 2nd edn. Sinauer Associates, Sunderland
Cornuet JM, Luikart G (1996) Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144:2001–2014
Cornuet JM, Santos F, Beaumont MA, Robert CP, Marin JM, Balding DJ, Guillemaud T, Estoup A (2008) Inferring population history with DIYABC: a user-friendly approach to approximate Bayesian computation. Bioinformatics 24:2713–2719
Cornuet JM, Pudlo P, Veyssier J, Dehne-Garcia A, Gautier M, Leblois R, Marin JM, Estoup A (2014) DIYABC v2.0: a software to make approximate Bayesian computation inferences about population history using single nucleotide polymorphism, DNA sequence and microsatellite data. Bioinformatics 30:1187–1189
Degner JF, Stout JL, Roth JD, Parkinson CL (2007) Population genetics and conservation of the threatened southeastern beach mouse (Peromyscus polionotus niveiventris): subspecies and evolutionary units. Conserv Genet 8:1441–1452
Duncan BW, Schmalzer PA (2004) Anthropogenic influences on potential fire spread in a pyrogenic ecosystem of Florida, USA. Landsc Ecol 19:153–165
Duncan BW, Boyle S, Breininger DR, Schmalzer PA (1999) Coupling past management practice and historic landscape change on John F. Kennedy Space Center, Florida. Landsc Ecol 14:291–309
Earl DA, von Holdt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Estoup A, Jarne P, Cornuet JM (2002) Homoplasy and mutation model at microsatellite loci and their consequences for population genetics analysis. Mol Ecol 11:1591–1604
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinform Online 1:47–50
Extine DD, Stout JI (1987) Dispersion and habitat occupancy of the beach mouse Peromyscus polionotus niveiventris. J Mammal 68:297–304
Fagundes NJR, Ray N, Beaumont MA, Neuenschwander S, Salzano F, Bonatto SL, Excoffier L (2007) Statistical evaluation of alternative models of human evolution. Proc Nat Acad Sci 104:17614–17619
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Forman RTT, Alexander LE (1998) Roads and their major ecological effects. Ann Rev Ecol Syst 29:207–231
Frankham R, Ballou J, Briscoe D (2009) Introduction to conservation genetics, 2nd edn. Cambridge Univ Press, Cambridge
Goldstein DB, Schlötterer C (1999) Microsatellites: evolution and applications. Oxford Univ Press, Oxford
Goudet J (2002) FSTAT, Version 2.9.3.2. Institute Ecol, Lausanne. http://www2.unil.ch/izea/softwares/fstat.html
Guillot G (2008) Inference of structure in subdivided populations at low levels of genetic differentiation—the correlated allele frequencies model revisited. Bioinformatics 24:2222–2228
Guillot G, Mortier F, Estoup A (2005) Geneland: a computer package for landscape genetics. Mol Ecol Res 5:712–715
Holliman DC (1983) Status and habitat of Alabama Gulf Coast beach mice Peromyscus polionotus ammobates and P. p. trisyllepsis. Northeast Gulf Sci 6:121–129
Humphrey SR, Kern WH, Ludlow MS (1987) Status survey of seven Florida mammals. Oxford Univ Press, Florida Coop Fish Wildl Res Unit Tech Rep #25
Kalkvik H, Stout J, Parkinson C (2012) Unraveling natural versus anthropogenic effects on genetic diversity within the southeastern beach mouse (Peromyscus polionotus niveiventris). Conserv Genet 13:1653–1664
Lynch M, Ritland K (1999) Estimation of pairwise relatedness with molecular markers. Genetics 152:1753–1766
McGregor RL, Bender DJ, Fahrig L (2008) Do small mammals avoid roads because of the traffic? J Appl Ecol 45:117–123
Mullen LM, Hirschmann RJ, Prince KL, Glenn TC, Dewey MJ, Hoekstra HE (2006) Sixty polymorphic microsatellite markers for the old field mouse developed in Peromyscus polionotus and Peromyscus maniculatus. Mol Ecol Notes 6:36–40
Neigel JE, Ball RM, Avise JC (1991) Estimation of single generation migration distances from geographic variation in animal mitochondrial DNA. Evolution 45:423–432
O’Brien S, Roelke M, Marker L, Newman A, Winkler C, Meltzer D et al (1985) Genetic basis for species vulnerability in the cheetah. Science 227:1428–1434
Oddy D, Hensley M, Provancha J, Smith R (1999) Long-distance dispersal of a southeastern beach mouse (Peromyscus polionotus niveiventris) at Cape Canaveral, Florida. Fla Field Nat 27:124–125
Oli M, Holler N, Wooten M (2001) Viability analysis of endangered Gulf Coast beach mice (Peromyscus polionotus) populations. Biol Conserv 97:107–118
Paetkau D, Slade R, Burdens M, Estoup A (2004) Genetic assignment methods for the direct, real-time estimation of migration rate: a simulation based exploration of accuracy and power. Mol Ecol 13:55–65
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Piry S, Luikart G, Cornuet JM (1999) Bottleneck: a computer program for detecting recent reductions in the effective population size using allele frequency data. J Hered 90:502–503
Piry S, Alapetite A, Cornuet JM, Paetkau D, Baudouin L, Estoup A (2004) GeneClass2: a software for genetic assignment and first-generation migrant detection. J Hered 95:536–539
Pries A, Branch L, Miller D (2009) Impact of hurricanes on habitat occupancy and spatial distribution of beach mice. J Mammal 90:841–850
Prince K, Glenn T, Dewey M (2002) Cross-species amplification among peromyscines of new microsatellite DNA loci from the oldfield mouse (Peromyscus polionotus subgriseus). Mol Ecol 2:133–136
Pritchard JK, Stephens M, Donnelly PJ (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Provancha JA, Oddy DM (1992) A mark and release study on the southeastern beach mouse (Peromyscus polionotus niveiventris) on the Kennedy Space Center, Florida. Fla Sci 1:28
R Core Team (2014) R: a language and environment for statistical computing. R Found Stat Comput, Vienna, http://www.R-project.org/
Rannala B, Mountain JL (1997) Detecting immigration by using multilocus genotypes. Proc Nat Acad Sci USA 94:9197–9201
Reed D, Frankham R (2003) Correlation between fitness and genetic diversity. Conserv Biol 17:230–237
Schielzeth H (2010) Simple means to improve the interpretability of regression coefficients. Meth Ecol Evol 1:103–113
Sikes RS, Gannon WL, Animal Care and Use Committee ASM (2011) Guidelines of the American society of mammalogists for the use of wild mammals in research. J Mammal 92:235–253
Sokal RR, Rohlf FJ (1995) Biometry, 3rd edn. WH Freeman Co, New York
Spielman D, Brook B, Frankham R (2004) Most species are not driven to extinction before genetic factors impact them. Proc Nat Acad Sci USA 101:15261–15264
Stout JI (1992) Southeastern beach mouse. In: Humphrey SR (ed) Rare and endangered biota of Florida, vol I., MammalsUniv Press Florida, Gainesville, pp 242–249
Suazo A, Fauth J, Roth J, Parkinson C, Stout J (2009) Responses of small rodents to habitat restoration and management for the imperiled Florida Scrub-Jay. Biol Conserv 142:2322–2328
Swilling WR, Wooten MC, Holler N, Lynn W (1998) Population dynamics of Alabama beach mice (Peromyscus polionotus ammobates) following hurricane Opal. Am Midl Nat 140:287–298
Underhill JE, Angold PG (1999) Effects of roads on wildlife in an intensively modified landscape. Environ Rev 8:21–39
Underwood JN, Smith LD, Van Oppen MJH, Gilmour JP (2007) Multiple scales of genetic connectivity in the brooding coral on isolated reefs following catastrophic bleaching. Mol Ecol 16:771–784
US Fish Wildlife Service (USFWS) (1993) Multi-species recovery plan for South Florida: southeastern beach mouse. http://www.fws.gov/verobeach/MSRPPDFs/SoutheasternBeachMouse.pdf
van Oosterhout C, Hutchinson B, Wills D, Shipley P (2004) MICROCHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Wooten M, Scribner K, Krehling J (1999) Isolation and characterization of microsatellite loci from the endangered beach mouse Peromyscus polionotus. Mol Ecol 8:157–168
Zuur AF, Ieno EN, Elphick CS (2010) A protocol for data exploration to avoid common statistical problems. Meth Ecol Evol 1:3–14
Acknowledgments
We thank NASA and Florida Institute of Technology for funding this research. We thank C. Hall and L. Phillips for aid in logistics and S. Gann, S. Legare, K. Holloway-Adkins, S. Weiss, S. Sneckenberger, and S. Trapp for tissue collection. We also thank M. Bush and J. Trefry for comments on an early version of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zimmerman, M., Oddy, D., Stolen, E. et al. Microspatial sampling reveals cryptic influences on gene flow in a threatened mammal. Conserv Genet 16, 1403–1414 (2015). https://doi.org/10.1007/s10592-015-0749-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-015-0749-6