Abstract
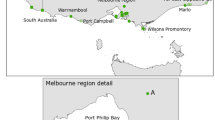
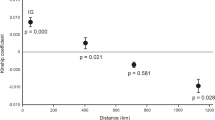
Habitat loss and fragmentation have important consequences for population persistence by reducing dispersal, connectivity, and population size. Characterizing patterns of population genetic structure and gene flow, and testing for the genetic signature of population declines and inbreeding can provide key insights into conservation for at-risk species. We use 12 microsatellite DNA markers to quantify the spatial structuring across two regional populations of the threatened eastern hog-nosed snake (Heterodon platirhinos) in Ontario, Canada. We further test for inbreeding and recent, rapid population declines, for the latter using both a ‘heterozygosity excess’ approach and Approximate Bayesian computation. Spatial Bayesian assignment tests revealed four distinct genetic clusters that were not apparent from species occurrence data, suggesting that the current designation of a single management unit for this species is invalid. We found some evidence for inbreeding in all studied locales. However, heterozygosity excess tests provided only weak evidence for recent bottlenecks and varied according to mutational model specified. In contrast, ABC provided strong support for a recent and rapid reduction in population size within one sample locale (Wasaga Beach), where individuals are largely confined to a small area circumscribed by expanding urban areas. For three other locales investigated, we found equal support for either recent rapid or gradual population declines. Immediate action is required for Wasaga Beach if this population is to persist.


Similar content being viewed by others
References
Allendorf FW (1986) Genetic drift and the loss of alleles versus heterozygosity. Zoo Biol 5:181–190
Beaumont MA, Zhang W, Balding DJ (2002) Approximate Bayesian computation in population genetics. Genetics 62:2025–2035
Benjamini Y, Yekutieli D (2001) The control of the false discovery rate in multiple testing under dependency. Ann Stat 29:1165–1188
Bertorelle G, Benazzo A, Mona S (2010) ABC as a flexible framework to estimate demography over space and time: some cons, many pros. Mol Ecol 19:2609–2625
Besag J, York J, Mollié A (1991) Bayesian image restoration, with two applications in spatial statistics. Ann Inst Stat Math 43:1–20
Bijlsma R, Bundgaard J, Boerema AC (2000) Does inbreeding affect the extinction risk of small populations? Predictions from Drosophila. J Evol Biol 12:1125–1137
Briskie JV, Mackintosh M (2004) Hatching failure increases with severity of population bottlenecks in birds. Proc Natl Acad Sci USA 101:558–561
Caccianiga M, Payette S (2006) recent advance of white spruce (Picea glauca) in the coastal tundra of the eastern shore of Hudson Bay (Québec, Canada). J Biogeogr 33:2120–2135
Chen C, Durand E, Forbes F, François O (2007) Bayesian clustering algorithms ascertaining spatial population structure: a new computer program and a comparison study. Mol Ecol Notes 7:747–756
Clark RW, Marchand MN, Clifford BJ, Stechert R, Stephens S (2011) Decline of an isolated timber rattlesnake (Crotalus horridus) population: interactions between climate change, disease, and loss of genetic diversity. Biol Conserv 144:886–891
Cornuet J-M, Santos F, Beaumont MA, Robert CP, Marin J-M, Balding DJ, Guillemaud T, Estoup A (2008) Inferring population history with DIY ABC: a user-friendly approach to approximate Bayesian computation. Bioinformatics 24:2713–2719
Cornuet J-M, Pudlo P, Veyssier J, Dehne-Garcia A, Gautier M, Leblois R, Marin J-M, Estoup A (2014) DIYABC v2.0: a software to make approximate Bayesian computation inferences about population history using single nucleotide polymorphism. DNA sequence and microsatellite data. Bioinformatics 30:1187–1189
COSEWIC (2007) COSEWIC assessment and update status report on the eastern hog-nosed snake Heterodon platirhinos in Canada. Committee on the Status of Endangered Wildlife in Canada, Ottawa
Coulon A, Guillot G, Cosson J-F, Angibault JMF, Aulagnier S, Cargnelutti B, Galan M, Hewison AJM (2006) Genetic structure is influenced by landscape features: empirical evidence from a roe deer population. Mol Ecol 15:1669–1679
Cunnington G (2004) Modeling habitat use by eastern hog-nosed snakes (Heterodon platirhinos) in Wasaga Beach Provincial Park. Thesis, Trent University, Peterborough, Ontario. B.Sc
Cunnington GM, Cebek JE (2005) Mating and nesting behavior of the eastern hognose snake (Heterodon platirhinos) in the northern portion of its range. Am Midl Nat 154:474–478
Durand E, Jay F, Gaggiotti OE, François O (2009) Spatial inference of admixture proportions and secondary contact zones. Mol Biol Evol 26:1963–1973
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Frankham R, Lees K, Montgomery ME, England PR, Lowe EH, Briscoe DA (1999) Do population size bottlenecks reduce evolutionary potential? Anim Conserv 2:255–260
Funk WC, Forsman ED, Johnson M, Mullins TD, Haig SM (2010) Evidence for recent population bottlenecks in northern spotted owls (Strix occidentalis caurina). Conserv Genet 11:1013–1021
Gamache I, Payette S (2005) Latitudinal response of subarctic tree lines to recent climate change in eastern Canada. J Biogeogr 32:849–862
Goudet J (2001) FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3). http://www.unil.ch/izea/softwares/fstat.html
Goudet J (2006) hierfstat: estimation and tests of hierarchical F-statistics. CRAN repository. http://cran.r-project.org/web/packages/hierfstat/
Green DM (2005) Designatable units for status assessment of endangered species. Conserv Biol 19(6):1813–1820
Harding JH (1997) Amphibians and reptiles of the Great Lakes region. University of Michigan Press, Ann Arbor
Hecnar SJ, M’Closkey RT (1998) Species richness patterns of amphibians in southwestern Ontario ponds. J Biogeogr 25:763–772
Hoffman JI, Grant SM, Forcada J, Phillips CD (2011) Bayesian inference of a historical bottleneck in a heavily exploited marine mammal. Mol Ecol 20:3989–4008
Ivanova NV, Dewaard JR, Hebert PDN (2006) An inexpensive, automation-friendly protocol for recovering high-quality DNA. Mol Ecol Notes 6:998–1002
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806
Jansen KP, Mushinsky HR, Karl SA (2008) Population genetics of the mangrove salt marsh snake, Nerodia clarkia compressicauda, in a linear, fragmented habitat. Conserv Genet 9:401–410
Keller LF, Waller DM (2002) Inbreeding effects in wild populations. Trends Ecol Evol 17:19–23
Lacy RC (1997) Importance of genetic variation to the viability of mammalian populations. J Mammal 78:320–335
Luikart G, Allendorf FW, Cornuet JM, Sherwin WB (1998) Distortion of allele frequency distributions provides a test for recent population bottlenecks. J Hered 89:238–247
Madsen T, Shine R, Olsen M, Wittzell H (1999) Conservation biology: restoration of an inbred adder population. Nature 402:34–35
Manel S, Bellemain E, Swenson JE, François O (2004) Assumed and inferred spatial structure of populations the Scandinavian brown bears revisited. Mol Ecol 13:1327–1331
Maruyama T, Fuerst PA (1985) Population bottlenecks and nonequilibrium models in population genetics. II. Number of alleles in a small population that was formed by a recent bottleneck. Genetics 111:675–689
McCoy RC, Garud NR, Kelley JL, Boggs CL, Petrov DA (2014) Genomic inference accurately predicts the timing and severity of a recent bottleneck in a nonmodel insect population. Mol Ecol 23:136–150
Moritz C (1994) Defining ‘Evolutionary Significant Units’ for conservation. Trends Ecol Evol 9:373–375
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Nei M, Maruyama T, Chakraborty R (1975) The bottleneck effect and genetic variability in populations. Evolution 29:1–10
Ontario Nature (2015) Ontario Reptile and Amphibian Atlas. http://www.ontarionature.org/protect/species/reptiles_and_amphibians/eastern_hognosed_snake.php
Palsbøll PJ, Bérubé M, Allendorf FW (2007) Identification of management units using population genetic data. Trends Ecol Evol 22:11–16
Peery MZ, Kirby R, Reid BN, Stoelting R, Doucet-Bëer E, Robinson S, Vásquez-Carillo C, Pauli JN, Palsbøll PJ (2012) Reliability of genetic bottleneck tests for detecting recent population declines. Mol Ecol 21:3403–3418
Piry S, Luikart G, Cornuet J-M (1999) BOTTLENECK: a computer program for detecting recent reductions in the effective population size using allele frequency data. J Hered 90:502–503
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Raymond M, Rousset F (1995) Genepop (version 1.2): population genetics software for exact tests and ecumenicism. Heredity 86:248–249
R Core Development Team (2012) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Robson L (2011) The spatial ecology of eastern hognose snakes (Heterodon platirhinos): habitat selection, home range size, and the effect of roads on movement patterns. MSc Thesis, University of Ottawa, Ottawa
Rosenberg NA (2004) DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes 4:137–138
Rouse JD, Willson RJ, Black R, Brooks RJ (2011) Movement and spatial dispersion of Sistrurus catenatus and Heterodon platirhinos: implications for interactions with roads. Copeia 2011:443–456
Row JR, Blouin-Demers G, Lougheed SC (2010) Habitat distribution influences dispersal and fine-scale genetic population structure of eastern foxsnakes (Mintonius gloydi) across a fragmented landscape. Mol Ecol 19:5157–5171
Row JR, Brooks RJ, MacKinnon CA, Lawson A, Crother BI, White M, Lougheed SC (2011) Approximate Bayesian computation reveals the factors that influence genetic diversity and population structure of foxsnakes. J Evol Biol 24:2364–2377
Saccheri IJ, Brakefield PM, Nichols RA (1996) Severe inbreeding depression and rapid fitness rebound in the butterfly Bicyclus anynana (Satyridae). Evolution 50:2000–2013
Schwartz TS, Karl SA (2005) Population and conservation genetics of the gopher tortoise (Gopherus polyphemus). Conserv Gen 6:917–928
Slatkin M (1995) A measure of population subdivision based on microsatellite allele frequencies. Genetics 139:457–462
Slatkin M (2008) Linkage disequilibrium—understanding the evolutionary past and mapping the medical future. Nat Rev Genet 9:477–485
Spencer CC, Neigel JE, Leberg PL (2000) Experimental evaluation of the usefulness of microsatellite DNA for detecting demographic bottlenecks. Mol Ecol 9:1517–1528
Spiegelhalter DJ, Best NG, Carlin BP, van der Linde A (2002) Bayesian measures of model complexity and fit. J R Statist Soc B 64:583–639
Storfer A, Mech SG, Reudink MW, Lew K (2014) Inbreeding and strong population subdivision in an endangered salamander. Conserv Genet 15:137–151
Thornton K, Andolfatto P (2006) Approximate Bayesian inference reveals evidence for a recent, severe bottleneck in a Netherlands population of Drosophila melanogaster. Genetics 172:1607–1619
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) Microchecker: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Wall WA, Douglas NA, Hoffmann WA, Wentworth TR, Gray JB, Xiang QJ, Knaus BK, Hohmann MG (2014) Evidence of population bottleneck in Astragalus michauxii (Fabaceae), a narrow endemic of the southeastern United States. Conserv Genet 15:153–164
Waples S (1995) Evolutionary Significant Units and the conservation of biological diversity under the Endangered Species Act. Am Fish Soc Symp 18:8–27
Williamson-Natesan EG (2005) Comparison of methods for detecting bottlenecks from microsatellite loci. Conserv Genet 6:551–562
Wright S (1951) The genetical structure of populations. Ann Eugen 15:323–354
Xuereb ATJ, Austin JD, Lougheed SC (2014) New microsatellite loci for the threatened eastern hog-nosed snake (Heterodon platirhinos) in Ontario, Canada. Conserv Genet Resour 6:69–71
Acknowledgments
We thank L. Robson, M. Gartshore, D. Jacobs, T. Tully, A. MacKenzie, K. Otterbein, T. Cameron, S. Sutton, R. Willson, G. Buck, R. Gould, C. Godwin, J. Feltham, T. Moore, G. Cundall, R. Hornsby, C. Jamison, and J. Quan for assistance with sample collection and Z. Sun for genotyping and J.D. Austin for microsatellite development. Funding was provided by NSERC (Discovery Grant to S.C.L. and CGS-M to A.T.J.X.) and the Species at Risk Research Fund for Ontario (SARRFO). We acknowledge support of Parks Canada, Ontario Parks, and Ontario Ministry of Natural Resources. Field collections were sanctioned by the Queen’s University Animal Care Committee (Lougheed-2008-059-R3), Endangered Species Act (PS-B-004-11), and Wildlife Scientific Collector’s Authorization (1062799, 1062623).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xuereb, A.T.J., Rouse, J.D., Cunnington, G. et al. Population genetic structure at the northern range limit of the threatened eastern hog-nosed snake (Heterodon platirhinos). Conserv Genet 16, 1265–1276 (2015). https://doi.org/10.1007/s10592-015-0737-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-015-0737-x