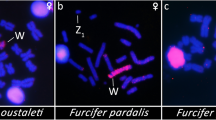
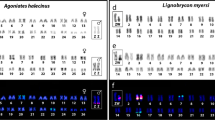
Abstract
Homomorphic sex chromosomes and their turnover are common in teleosts. We investigated the evolution of nascent sex chromosomes in several populations of two sister species of African annual killifishes, Nothobranchius furzeri and N. kadleci, focusing on their under-studied repetitive landscape. We combined bioinformatic analyses of the repeatome with molecular cytogenetic techniques, including comparative genomic hybridization, fluorescence in situ hybridization with satellite sequences, ribosomal RNA genes (rDNA) and bacterial artificial chromosomes (BACs), and immunostaining of SYCP3 and MLH1 proteins to mark lateral elements of synaptonemal complexes and recombination sites, respectively. Both species share the same heteromorphic XY sex chromosome system, which thus evolved prior to their divergence. This was corroborated by sequence analysis of a putative master sex determining (MSD) gene gdf6Y in both species. Based on their divergence, differentiation of the XY sex chromosome pair started approximately 2 million years ago. In all populations, the gdf6Y gene mapped within a region rich in satellite DNA on the Y chromosome long arms. Despite their heteromorphism, X and Y chromosomes mostly pair regularly in meiosis, implying synaptic adjustment. In N. kadleci, Y-linked paracentric inversions like those previously reported in N. furzeri were detected. An inversion involving the MSD gene may suppress occasional recombination in the region, which we otherwise evidenced in the N. furzeri population MZCS-121 of the Limpopo clade lacking this inversion. Y chromosome centromeric repeats were reduced compared with the X chromosome and autosomes, which points to a role of relaxed meiotic drive in shaping the Y chromosome repeat landscape. We speculate that the recombination rate between sex chromosomes was reduced due to heterochiasmy. The observed differences between the repeat accumulations on the X and Y chromosomes probably result from high repeat turnover and may not relate closely to the divergence inferred from earlier SNP analyses.






Similar content being viewed by others
Data availability
The following nucleotide sequences were deposited in NCBI GenBank: 5S rDNA from N. kadleci (accession number MZ694985), 18S rDNA from N. guentheri (MZ682111), and X- and Y-linked gdf6 gene variants from N. kadleci (ON921000-ON921004). Measurements related to the construction of cytogenetic maps of sex chromosomes are available from the Dryad digital data repository: https://doi.org/10.5061/dryad.4mw6m90ck (Štundlová et al. 2022). All other relevant data are within the paper and its supplementary material.
Abbreviations
- 2n:
-
Diploid chromosome number
- a:
-
Acrocentric chromosome
- BAC:
-
Bacterial artificial chromosome
- BSA:
-
Bovine serum albumin
- CDS:
-
Coding sequence
- CGH:
-
Comparative genomic hybridization
- CMA3 :
-
Chromomycin A3
- DABCO:
-
1,4-Diazabicyclo(2.2.2)-octane
- DAPI:
-
4′,6-Diamidino-2-phenylindole
- DOP-PCR:
-
Degenerate oligonucleotide-primed PCR
- dUTP:
-
2′-Deoxyuridine-5′-triphosphate
- FISH:
-
Fluorescence in situ hybridization
- FITC:
-
Fluorescein isothiocyanate
- gdf6 :
-
Growth differentiation factor 6
- gDNA:
-
Genomic DNA
- KY:
-
Thousand years
- m:
-
Metacentric chromosome
- MLH1:
-
MutL homolog 1
- MSD:
-
Master sex-determining (gene)
- MYA:
-
Million years ago
- MY:
-
Million years
- ND-FISH:
-
Non-denaturing FISH
- NGS:
-
Normal goat serum
- p-arm:
-
Short chromosome arm
- PBS:
-
Phosphate-buffered saline
- PCR:
-
Polymerase chain reaction
- q-arm:
-
Long chromosome arm
- rDNA:
-
Ribosomal DNA
- RT:
-
Room temperature
- SA:
-
Sexual antagonism
- SD:
-
Sex-determining (region)
- SDS:
-
Sodium dodecyl sulfate
- SNP:
-
Single-nucleotide polymorphism
- SSC:
-
Saline-sodium citrate
- SYCP3:
-
Synaptonemal complex protein 3
- st:
-
Subtelocentric chromosome
- UTR:
-
Untranslated region
References
Akera T, Trimm E, Lampson MA (2019) Molecular strategies of meiotic cheating by selfish centromeres. Cell 178:1132–1144. https://doi.org/10.1016/j.cell.2019.07.001
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic Local Alignment Search Tool. J Mol Biol 215:403–410. http://blast.ncbi.nlm.nih.gov/blast
Andrews S (2010) FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/. Accessed 25 Sept 2019
Araya-Jaime C, Serrano ÉA, de Andrade Silva DMZ, Yamashita M, Iwai T, Oliveira C, Foresti F (2015) Surface-spreading technique of meiotic cells and immunodetection of synaptonemal complex proteins in teleostean fishes. Mol Cytogenet 8:4. https://doi.org/10.1186/s13039-015-0108-9
Bachtrog D, Mank JE, Peichel CL, Kirkpatrick M, Otto SP, Ashman TL, Hahn MW, Kitano J, Mayrose I, Ming R et al (2014) Sex determination: why so many ways of doing it? PLoS Biol 12:e1001899. https://doi.org/10.1371/journal.pbio.1001899
Bartáková V, Reichard M, Janko K, Polačik M, Blažek R, Reichwald K, Cellerino A, Bryja J (2013) Strong population genetic structuring in an annual fish, Nothobranchius furzeri, suggests multiple savannah refugia in southern Mozambique. BMC Evol Biol 13:196. https://doi.org/10.1186/1471-2148-13-196
Bartáková V, Reichard M, Blažek R, Polačik M, Bryja J (2015) Terrestrial fishes: rivers are barriers to gene flow in annual fishes from the African savanna. J Biogeogr 42:1832–1844. https://doi.org/10.1111/jbi.12567
Bergero R, Charlesworth D (2009) The evolution of restricted recombination in sex chromosomes. Trends Ecol Evol 24:94–102. https://doi.org/10.1016/j.tree.2008.09.010
Bergero R, Gardner J, Bader B, Yong L, Charlesworth D (2019) Exaggerated heterochiasmy in a fish with sex-linked male coloration polymorphisms. Proc Natl Acad Sci U S A 116:6924–6931. https://doi.org/10.1073/pnas.1818486116
Bertollo LAC, Cioffi MB, Moreira-Filho O (2015) Direct chromosome preparation from freshwater teleost fishes. In: Ozouf-Costaz C, Pisano E, Foresti F, and de Almeida-Toledo LF (eds) Fish cytogenetic techniques: ray-fin fishes and chondrichthyans. CRC Press, Inc, Endfield, pp 21–26. https://doi.org/10.1201/b18534-4
Blažek R, Polačik M, Kačer P, Cellerino A, Řežucha R, Methling C, Tomášek O, Syslová K, Terzibasi Tozzini E, Albrecht T, Vrtílek M, Reichard M (2017) Repeated intraspecific divergence in life span and aging of African annual fishes along an aridity gradient. Evolution 2017;71: 386–402. https://doi.org/10.1111/evo.13127
Cellerino A, Valenzano DR, Reichard M (2016) From the bush to the bench: the annual Nothobranchius fishes as a new model system in biology. Biol Rev Camb Philos Soc 91:511–533. https://doi.org/10.1111/brv.12183
Chalopin D, Volff J-N, Galiana D, Anderson JL, Schartl M (2015) Transposable elements and early evolution of sex chromosomes in fish. Chromosome Res 23:545–560. https://doi.org/10.1007/s10577-015-9490-8
Charlesworth D (2018) The guppy sex chromosome system and the sexually antagonistic polymorphism hypothesis for Y chromosome recombination suppression. Genes 9:264. https://doi.org/10.3390/genes9050264
Charlesworth D (2019) Young sex chromosomes in plants and animals. New Phytol 224:1095–1107. https://doi.org/10.1111/nph.16002
Charlesworth D (2021) When and how do sex-linked regions become sex chromosomes? Evolution 75:569–581. https://doi.org/10.1111/evo.14196
Charlesworth B, Charlesworth D (1978) A model for the evolution of dioecy and gynodioecy. Am Nat 112:975–997. https://doi.org/10.1086/283342
Charlesworth B, Sniegowski P, Stephan W (1994) The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371:215–220. https://doi.org/10.1038/371215a0
Charlesworth D, Charlesworth B, Marais G (2005) Steps in the evolution of heteromorphic sex chromosomes. Heredity 95:118–128. https://doi.org/10.1038/sj.hdy.6800697
Chen J, Fu Y, Xiang D, Zhao G, Long H, Liu J, Yu Q (2008) XX/XY heteromorphic sex chromosome systems in two bullhead catfish species, Liobagrus marginatus and L. styani (Amblycipitidae, Siluriformes). Cytogenet Genome Res 122:169–174. https://doi.org/10.1159/000163095
Cioffi MB, Martins C, Vicari MR, Rebordinos L, Bertollo LAC (2010) Differentiation of the XY sex chromosomes in the fish Hoplias malabaricus (Characiformes, Erythrinidae). Unusual accumulation of repetitive sequences on the X chromosome. Sex Dev 4:176–185. https://doi.org/10.1159/000309726
Cioffi MB, Liehr T, Trifonov V, Molina WF, Bertollo LAC (2013) Independent sex chromosome evolution in lower vertebrates: a molecular cytogenetic overview in the Erythrinidae fish family. Cytogenet Genome Res 141:186–194. https://doi.org/10.1159/000354039
Cuadrado A, Jouve N (2010) Chromosomal detection of simple sequence repeats (SSRs) using nondenaturing FISH (ND-FISH). Chromosoma 119:495–503. https://doi.org/10.1007/s00412-010-0273-x
Cui R, Medeiros T, Willemsen D, Iasi LNM, Collier GE, Graef M, Reichard M, Valenzano DR (2019) Relaxed selection limits lifespan by increasing mutation load. Cell 178:385-399.e20. https://doi.org/10.1016/j.cell.2019.06.004
de Almeida-Toledo LF, Foresti F, Péquignot EV, Daniel-Silva MFZ (2001) XX:XY sex chromosome system with X heterochromatinization: an early stage of sex chromosome differentiation in the Neotropic electric eel Eigenmannia virescens. Cytogenet Cell Genet 95:73–78. https://doi.org/10.1159/000057020
Dorn A, Musilová Z, Platzer M, Reichwald K, Cellerino A (2014) The strange case of East African annual fishes: aridification correlates with diversification for a savannah aquatic group? BMC Evol Biol 14:1–13. https://doi.org/10.1186/s12862-014-0210-3
Dorn A, Ng’oma E, Janko K, Reichwald K, Polačik M, Platzer M, Cellerino A, Reichard M (2011) Phylogeny, genetic variability and colour polymorphism of an emerging animal model: the short-lived annual Nothobranchius fishes from southern Mozambique. Mol Phylogenet Evol 61:739–749. https://doi.org/10.1016/j.ympev.2011.06.010
El Taher A, Ronco F, Matschiner M, Salzburger W, Böhne A (2021) Dynamics of sex chromosome evolution in a rapid radiation of cichlid fishes. Sci Adv 7:eabe8215 . https://doi.org/10.1126/sciadv.abe8215
Ewulonu UK, Haas R, Turner B (1985) A multiple sex chromosome system in the annual killfish, Nothobranchiu guentheri. Copeia 2:503–508. https://doi.org/10.2307/1444868
Flynn JM, Caldas I, Cristescu ME, Clark AG (2017) Selection constrains high rates of tandem repetitive DNA mutation in Daphnia pulex. Genetics 207:697–710. https://doi.org/10.1534/genetics.117.300146
Flynn JM, Lower SE, Barbash DA, Clark AG (2018) Rates and patterns of mutation in tandem repetitive DNA in six independent lineages of Chlamydomonas reinhardtii. Genome Biol Evol 10:1673–1686. https://doi.org/10.1093/gbe/evy123
Fricke R, Eschmeyer WN, Van der Laan R (eds) (2022) Eschmeyer’s catalog of fishes: genera, species, references. http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp. Accessed 11 March 2022
Furman BLS, Metzger DCH, Darolti I, Wright AE, Sandkam BA, Almeida P, Shu JJ, Mank JE (2020) Sex chromosome evolution: so many exceptions to the rules. Genome Biol Evol 12:750–763. https://doi.org/10.1093/gbe/evaa081
Garrido-Ramos MA (2017) Satellite DNA: an evolving topic. Genes 8:230. https://doi.org/10.3390/genes8090230
Glugoski L, Deon G, Schott S, Vicari MR, Nogaroto V, Moreira-Filho O (2020) Comparative cytogenetic analyses in Ancistrus species (Siluriformes: Loricariidae). Neotrop Ichthyol 18:e200013. https://doi.org/10.1590/1982-0224-2020-0013
Guiguen Y, Fostier A, Herpin A (2019) Sex determination and differentiation in fish: genetic, genomic, and endocrine aspects. In: Wang HP, Piferrer F, Chen S-L (eds) Sex Control in Aquaculture, 1st edn. John Wiley & Sons, Hoboken, pp 35–63
Haaf T, Schmid M (1984) An early stage of ZZ/ZW sex chromosomes differentiation in Poecilia sphenops var. melanistica (Poeciliidae, Cyprinodontiformes). Chromosoma 89:37–41. https://doi.org/10.1007/BF00302348
Hu CK, Brunet A (2018) The African turquoise killifish: a research organism to study vertebrate aging and diapause. Aging Cell 17:1–15. https://doi.org/10.1111/acel.12757
Iwata-Otsubo A, Dawicki-McKenna JM, Akera T, Falk SJ, Chmátal L, Yang K, Sullivan BA, Schultz RM, Lampson MA, Black BE (2017) Expanded satellite repeats amplify a discrete CENP-A nucleosome assembly site on chromosomes that drive in female meiosis. Curr Biol 27:2365–2373.e8. https://doi.org/10.1016/j.cub.2017.06.069
Jay P, Tezenas E, Véber A, Giraud T (2022) Sheltering of deleterious mutations explains the stepwise extension of recombination suppression on sex chromosomes and other supergenes. PloS Biol 20:e3001698. https://doi.org/10.1371/journal.pbio.3001698
Jeffries DL, Gerchen JF, Scharmann M, Pannell J (2021) A neutral model for the loss of recombination on sex chromosomes. Phil Trans R Soc B 376:20200096. https://doi.org/10.1098/rstb.2020.0096
Johnson Pokorná M, Reifová R (2021) Evolution of B chromosomes: from dispensable parasitic chromosomes to essential genomic players. Front Genet 12:727570. https://doi.org/10.3389/fgene.2021.727570
Kato A, Albert P, Vegy J, Bircher J (2006) Sensitive fluorescence in situ hybridization signal detection in maize using directly labelled probes produced by high concentration DNA polymerase nick translation. Boitech Histochem 81:71–78. https://doi.org/10.1080/10520290600643677
Kent TV, Uzunović J, Wright SI (2017) Coevolution between transposable elements and recombination. Philos Trans R Soc B Biol Sci 372:20160458. https://doi.org/10.1098/rstb.2016.0458
King M (1993) Species evolution: the role of chromosome change. Cambridge University Press, Cambridge
Kitano J, Ross JA, Mori S, Kume M, Jones FC, Chan YF, Absher DM, Grimwood J, Schmutz J, Myers RM et al (2009) A role for a neo-sex chromosome in stickleback speciation. Nature 461:1079–1083. https://doi.org/10.1038/nature08441
Kratochvíl L, Stöck M, Rovatsos M, Bullejos M, Herpin A, Jeffries DL, Peichel CL, Perrin N, Valenzuela N, Johnson Pokorná M (2021) Expanding the classical paradigm: what we have learnt from vertebrates about sex chromosome evolution. Phil Trans R Soc B 376:20200097. https://doi.org/10.1098/rstb.2020.0097
Krysanov E, Demidova T (2018) Extensive karyotype variability of African fish genus Nothobranchius (Cyprinodontiformes). Comp Cytogenet 12:387–402. https://doi.org/10.3897/CompCytogen.v12i3.25092
Krysanov E, Demidova T, Nagy B (2016) Divergent karyotypes of the annual killifish genus Nothobranchius (Cyprinodontiformes, Nothobranchiidae). Comp Cytogenet 10:439–445. https://doi.org/10.3897/CompCytogen.v10i3.9863
Kursel LE, Malik HS (2018) The cellular mechanisms and consequences of centromere drive. Curr Opin Cell Biol 52:58–65. https://doi.org/10.1016/j.ceb.2018.01.011
Lenormand T, Roze D (2022) Y recombination arrest and degeneration in the absence of sexual dimorphism. Science 375:663–666. https://doi.org/10.1126/science.abj1813
Levan AK, Fredga K, Sandberg AA (1964) Nomenclature for centromeric position on chromosomes. Hereditas 52:201–220. https://doi.org/10.1111/j.1601-5223.1964.tb01953.x
Li S-F, Su T, Cheng G-Q, Wang B-X, Li X, Deng C-L, Gao W-J (2017) Chromosome evolution in connection with repetitive sequences and epigenetics in plants. Genes 8:290. https://doi.org/10.3390/genes8100290
Lisachov AP, Zadesenets KS, Rubtsov NB, Borodin PM (2015) Sex chromosome synapsis and recombination in male guppies. Zebrafish 12:174–180. https://doi.org/10.1089/zeb.2014.1000
Mank JE, Avise JC (2009) Evolutionary diversity and turn-over of sex determination in teleost fishes. Sex Dev 3:60–67. https://doi.org/10.1159/000223071
Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10–12. https://doi.org/10.14806/ej.17.1.200
Martins C, Wasko AP (2004) Organization and evolution of 5S ribosomal DNA in the fish genome. In: Williams CR (ed) Focus on Genome Research. Nova Science Publishers, Hauppauge, pp 335–363
Mayr B, Ráb P, Kalat M (1985) Localisation of NORs and counterstain-enhanced fluorescence studies in Perca fluviatilis (Pisces, Percidae). Genetica 67:51–56. https://doi.org/10.1007/BF02424460
Moens PB (2006) Zebrafish: chiasmata and interference. Genome 49:205–208. https://doi.org/10.1139/g06-021
Nanda I, Schories S, Simeonov I, Adolfi MC, Du K, Steinlein C, Alsheimer M, Haaf T, Schartl M (2022) Evolution of the degenerated Y-chromosome of the swamp guppy, Micropoecilia picta. Cells 11:1118. https://doi.org/10.3390/cells11071118
Nath S, Welch LA, Flanagan MK, White MA (2022) Meiotic pairing and double-strand break formation along the heteromorphic threespine stickleback sex chromosomes. Chromosome Res. https://doi.org/10.1007/s10577-022-09699-0
Natri HM, Merilä J, Shikano T (2019) The evolution of sex determination associated with a chromosomal inversion. Nat Commun 10:145. https://doi.org/10.1038/s41467-018-08014-y
Navrátilová A, Koblížková A, Macas J (2008) Survey of extrachromosomal circular DNA derived from plant satellite repeats. BMC Plant Biol 8:90. https://doi.org/10.1186/1471-2229-8-90
Ng’oma E, Groth M, Ripa R, Platzer M, Cellerino A (2014) Transcriptome profiling of natural dichromatism in the annual fishes Nothobranchius furzeri and Nothobranchius kadleci. BMC Genomics 15:754. https://doi.org/10.1186/1471-2164-15-754
Novák P, Neumann P, Macas J (2020) Global analysis of repetitive DNA from unassembled sequence reads using RepeatExplorer2. Nat Protoc 15:3745–3776. https://doi.org/10.1038/s41596-020-0400-y
Palmer DH, Rogers TF, Dean R, Wright AE (2019) How to identify sex chromosomes and their turnover. Mol Ecol 28:4709–4724. https://doi.org/10.1111/mec.15245
Pan Q, Feron R, Jouanno E, Darras H, Herpin A, Koop B, Rondeau E, Goetz FW, Larson WA, Bernatchez L et al (2021) The rise and fall of the ancient northern pike master sex determining gene. Elife 10:e62858. https://doi.org/10.7554/eLife.62858
Peichel C, McCann SR, Ross JA, Naftaly AFS, Urton JR, Cech JN, Grimwood J, Schmutz J, Myers RM, Kingsley DM, White MA (2020) Assembly of a young vertebrate Y chromosome reveals convergent signatures of sex chromosome evolution. Genome Biol 21:177. https://doi.org/10.1186/s13059-020-02097-x
Peona V, Blom MPK, Xu L, Burri R, Sullivan S, Bunikis I, Liachko I, Haryoko T, Jønsson KA, Zhou Q, Irestedt M, Suh A (2020) Identifying the causes and consequences of assembly gaps using a multiplatform genome assembly of a bird-of-paradise. Mol Ecol Resour 21:263–286. https://doi.org/10.1111/1755-0998.13252
Perrin N (2021) Sex-chromosome evolution in frogs: what role for sex-antagonistic genes? Phil Trans R Soc B 376:20200094. https://doi.org/10.1098/rstb.2020.0094
Pokorná M, Altmanová M, Kratochvíl L (2014) Multiple sex chromosomes in the light of female meiotic drive in amniote vertebrates. Chromosome Res 22:35–44. https://doi.org/10.1007/s10577-014-9403-2
Ráb P, Roth P (1988) Cold-blooded vertebrates. In: Balicek P, Forejt J, Rubeš J (eds) Methods of chromosome analysis. Cytogenetická Sekce Československé Biologické Společnosti při CSAV, Brno, Czech Republic, pp 115–124
Reichard M, Giannetti K, Ferreira T, Maouche A, Vrtílek M, Polačik M, Blažek R, Ferreira MG (2021) Lifespan and telomere length variation across populations of wild-derived African killifish. Mol Ecol. https://doi.org/10.1111/mec.16287
Reichwald K, Lauber C, Nanda I, Kirschner J, Hartmann N, Schories S, Gausmann U, Taudien S, Schilhabel MB, Szafranski K et al (2009) High tandem repeat content in the genome of the short-lived annual fish Nothobranchius furzeri: a new vertebrate model for aging research. Genome Biol 10:R16. https://doi.org/10.1186/gb-2009-10-2-r16
Reichwald K, Petzold A, Koch P, Downie BR, Hartmann N, Pietsch S, Baumgart M, Chalopin D, Felder M, Bens M et al (2015) Insights into sex chromosome evolution and aging from the genome of a short-lived fish. Cell 163:1527–1538. https://doi.org/10.1016/j.cell.2015.10.071
Richter A, Krug J, Englert C (2022) Molecular sexing of Nothobranchius furzeri embryos and larvae. Cold Spring Harb Protoc. https://doi.org/10.1101/pdb.prot.107782
Rifkin JL, Beaudry FEG, Humphries Z, Choudhury BI, Barrett SCH, Wright SI (2021) Widespread recombination suppression facilitates plant sex chromosome evolution. Mol Biol Evol 38:1018–1030. https://doi.org/10.1093/molbev/msaa271
Sakamoto Y, Zacaro AA (2009) LEVAN, an ImageJ plugin for morphological cytogenetic analysis of mitotic and meiotic chromosomes. Initial version. An open source Java plugin distributed over the Internet from http://rsbweb.nih.gov/ij/. Accessed 10 Jan 2022
Sardell JM, Cheng C, Dagilis AJ, Ishikawa A, Kitano J, Peichel CL, Kirkpatrick M (2018) Sex differences in recombination in sticklebacks. G3 (Bethesda) 8:1971–1983. https://doi.org/10.1534/g3.118.200166
Schartl M, Schmid M, Nanda I (2016) Dynamics of vertebrate sex chromosome evolution: from equal size to giants and dwarfs. Chromosoma 125:553–571. https://doi.org/10.1007/s00412-015-0569-y
Sember A, Bohlen J, Šlechtová V, Altmanová M, Symonová R, Ráb P (2015) Karyotype differentiation in 19 species of river loach fishes (Nemacheilidae, Teleostei): extensive variability associated with rDNA and heterochromatin distribution and its phylogenetic and ecological interpretation. BMC Evol Biol 15:251. https://doi.org/10.1186/s12862-015-0532-9
Sember A, Bertollo LAC, Ráb P, Yano CF, Hatanaka T, de Oliveira EA, Cioffi MB (2018a) Sex chromosome evolution and genomic divergence in the fish Hoplias malabaricus (Characiformes, Erythrinidae). Front Genet 9:3389. https://doi.org/10.3389/fgene.2018.00071
Sember A, Bohlen J, Šlechtová V, Altmanová M, Pelikánová Š, Ráb P (2018b) Dynamics of tandemly repeated DNA sequences during evolution of diploid and tetraploid botiid loaches (Teleostei: Cobitoidea: Botiidae). PLoS One 13:1–27. https://doi.org/10.1371/journal.pone.0195054
Sember A, Nguyen P, Perez MF, Altmanová M, Ráb P, Cioffi MB (2021) Multiple sex chromosomes in teleost fishes from a cytogenetic perspective: state of the art and future challenges. Phil Trans R Soc B 376:20200098. https://doi.org/10.1098/rstb.2020.0098
Sember A, Pelikánová Š, Cioffi M de B, Šlechtová V, Hatanaka T, Do Doan H, Knytl M, Ráb P (2020) Taxonomic diversity not associated with gross karyotype differentiation: the case of bighead carps, genus Hypophthalmichthys (Teleostei, Cypriniformes, Xenocyprididae). Genes 11:479. https://doi.org/10.3390/genes11050479
Smit AFA, Hubley R, Green P (2013–2015) RepeatMasker Open-4.0. http://www.repeatmasker.org. Accessed 2 Jan 2022
Sola L, Rossi AR, Iaselli V, Rasch EM, Monaco PJ (1992) Cytogenetics of bisexual/unisexual species of Poecilia. II. Analysis of heterochromatin and nucleolar organizer regions in Poecilia mexicana mexicana by C-banding and DAPI, quinacrine, chromomycin A3, and silver staining. Cytogenet Cell Genet 60:229–235. https://doi.org/10.1159/000133346
Stephan W (1986) Recombination and the evolution of satellite DNA. Genet Res 47:167–174. https://doi.org/10.1017/s0016672300023089
Štundlová J, Kreklová M, Lukšíková K, Voleníková A, Pavlica T, Altmanová M, Reichard M, Dalíková M, Pelikánová Š, Marta A, et al. (2022) Data from: Sex chromosome differentiation via changes in the Y chromosome repeat landscape in African annual killifishes Nothobranchius furzeri and N. kadleci, Dryad Dataset. https://doi.org/10.5061/dryad.4mw6m90ck
Sutherland BJG, Rico C, Audet C, Bernatchez L (2017) Sex chromosome evolution, heterochiasmy, and physiological QTL in the salmonid brook charr Salvelinus fontinalis. G3 (Bethesda) 7:2749–2762. https://doi.org/10.1534/g3.117.040915
Torgasheva AA, Malinovskaya LP, Zadesenets KS, Karamysheva TV, Kizilova EA, Akberdina EA, Pristyazhnyuka IE, Shnaiderc EP, Volodkinad VA, Saifitdinovad AF et al (2019) Germline-restricted chromosome (GRC) is widespread among songbirds. Proc Nat Acad Sci 116:11845–11850. https://doi.org/10.1073/pnas.1817373116
Torgasheva A, Malinovskaya L, Zadesenets KS, Slobodchikova A, Shnaider E, Rubtsov N, Borodin P (2021) Highly conservative pattern of sex chromosome synapsis and recombination in Neognathae birds. Genes 12:1358. https://doi.org/10.3390/genes12091358
Traut W, Winking H (2001) Meiotic chromosomes and stages of sex chromosome evolution in fish: zebrafish, platyfish and guppy. Chromosome Res 9:659–672. https://doi.org/10.1023/A:1012956324417
Valenzano DR, Benayoun BA, Singh PP, Zhang E, Etter PD, Hu CK, Clément-Ziza M, Willemsen D, Cui R, Harel I et al (2015) The African turquoise killifish genome provides insights into evolution and genetic architecture of lifespan. Cell 163:1539–1554. https://doi.org/10.1016/j.cell.2015.11.008
van der Merwe PDW, Cotterill FPD, Kandziora M, Watters BR, Nagy B, Genade T, Flügel TJ, Svendsen DS, Bellstedt DU (2021) Genomic fingerprints of palaeogeographic history: the tempo and mode of rift tectonics across tropical Africa has shaped the diversification of the killifish genus Nothobranchius (Teleostei: Cyprinodontiformes). Mol Phylogenet Evol 158:106988. https://doi.org/10.1016/j.ympev.2020.106988
Völker M, Ráb P (2015) Direct chromosome preparation from regenerating fin tissue. In: Ozouf-Costaz C, Pisano E, Foresti F, and de Almeida-Toledo LF (eds) Fish cytogenetic techniques: ray-fin fishes and chondrichthyans. CRC Press, Inc, Endfield, pp 37–41 . https://doi.org/10.1201/b18534-4
Wichman HA, Payne CT, Ryder OA, Hamilton MJ, Maltbie M, Baker RJ (1991) Genomic distribution of heterochromatic sequences in equids: implications to rapid chromosomal evolution. J Hered 82:369–377. https://doi.org/10.1093/oxfordjournals.jhered.a111106
Wildekamp RH (2004) A world of killies: atlas of the oviparous cyprinodontiform fishes of the world (Vol. 4). The American Killifish Association, Elyria
Willemsen D, Cui R, Reichard M, Valenzano DR (2020) Intra-species differences in population size shape life history and genome evolution. Elife 9:e55794. https://doi.org/10.7554/eLife.55794
Yang F, Trifonov V, Ng BL, Kosyakova N, Carter NP (2009) In: Liehr T (ed) Generation of paint probes by flow-sorted and microdissected chromosomes. Fluorescence in situ hybridization (FISH)—application guide, 2nd edn. Springer, Berlin, pp 35–52
Yano CF, Bertollo LAC, Ezaz T, Trifonov V, Sember A, Liehr T, Cioffi MB (2017) Highly conserved Z and molecularly diverged W chromosomes in the fish genus Triportheus (Characiformes, Triportheidae). Heredity 118:276–283. https://doi.org/10.1038/hdy.2016.83F
Yano CF, Sember A, Kretschmer R, Bertollo LAC, Ezaz T, Hatanaka T, Liehr T, Ráb P, Al-Rikabi A, Viana PF et al (2021) Against the mainstream: exceptional evolutionary stability of ZW sex chromosomes across the fish families Triportheidae and Gasteropelecidae (Teleostei: Characiformes). Chromosome Res 29:391–416. https://doi.org/10.1007/s10577-021-09674-1
Yoshida K, Kitano J (2012) The contribution of female meiotic drive to the evolution of neo-sex chromosomes. Evolution 66:3198–3208. https://doi.org/10.1111/j.1558-5646.2012.01681.x
Yoshido A, Sahara K, Yasukochi Y. Silk moths (Lepidoptera) (2015) In: Sharakhov IV (ed) Protocols for cytogenetic mapping of arthropod genomes. CRC Press, Boca Raton, FL, USA, pp 219–256
Acknowledgements
The authors are grateful to S. Förste and M. Platzer (The Leibniz Institute on Aging – Fritz Lipmann Institute, Jena, Germany) for providing BAC clones from the N. furzeri genomic library, to D. R. Valenzano and D. E. M. de Bakker (The Leibniz Institute on Aging – Fritz Lipmann Institute, Jena, Germany) for providing fish material for the gdf6 sequence analyses and P. Koch (Core Facility Life Science Computing, The Leibniz Institute on Aging – Fritz Lipmann Institute, Jena, Germany) for providing alignments of genomic sequencing reads from Reichwald et al. (2015). We are also thankful to P. Šejnohová and D. Dedukh (both Institute of Animal Physiology and Genetics, Czech Academy of Sciences, Liběchov, Czech Republic) for the laboratory assistance and advice on the immunostaining protocol, respectively, and to C. Johnson as well as A.-M. Dion-Côté (Université de Moncton, Canada) for copyediting. We would also like to thank two anonymous reviewers for their feedback and valuable comments.
Funding
This study was supported by The Czech Science Foundation (grant no. 19-22346Y) (J. Š., P. N., M. H., K. L., A. V., T. P., M. A., Š. P., M. H., M. J., A. S.) and further by a grant (EN 280/15–1) from the Deutsche Forschungsgemeinschaft (C. E., A. R.) and the projects EXCELLENCE CZ.02.1.01/0.0/0.0/15_003/0000460 OP RDE (Ministry of Education, Youth and Sports) (P. R.); RVO:67985904 of IAPG CAS, Liběchov (Czech Academy of Sciences) (M. A., A. M., T. D., J. B., Š. P., P. R., A. S.); and the Charles University Research Centre program No. 204069 (M. A.). Computational resources were supplied by the project “e-Infrastruktura CZ” (e-INFRA LM2018140) provided within the program Projects of Large Research, Development and Innovations Infrastructures and the ELIXIR-CZ project (LM2018131), part of the international ELIXIR infrastructure. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
Conceptualization: A. S., P. N., M. R.; data curation: P. N., A. V., M. D., T. D., A. R.; formal analysis: J. Š., P. N., A. V., M. D.; funding acquisition: A. S., C. E.; investigation: J. Š., M. H., K. L., A. V., T. P., M. A., A. R., M. D., Š. P., A. M., S. A. S., M. H., M. J., T. D., J. B., P. N., A. S.; methodology: J. Š., A. S., P. N., A. V., M. D., A. M., S. A. S., A. R.; project administration: A. S., P. N.; resources: A. S., P. N., M. R., C. E., P. R.; supervision: A. S., P. N., M. R., P. R., J. B.; validation: J. Š., A. S., P. N., A. R.; writing original draft: A. S.; writing—review and editing: P. N., A. S., A. R., J. Š., M. R., A. V., M. A., J. B., P. R., S. A. S., M. D., C. E.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
To prevent fish suffering, all handling of fish individuals followed European standards in agreement with §17 of the Act No. 246/1992 coll. The procedures involving fishes were supervised by the Institutional Animal Care and Use Committee of the Institute of Animal Physiology and Genetics CAS, v.v.i., and the supervisor’s permit number CZ 02361 was certified and issued by the Ministry of Agriculture of the Czech Republic. For direct preparations of chromosomes from kidney and gonads, fishes were euthanized using 2-phenoxyethanol (Sigma-Aldrich) before organ sampling. Fin samples (a narrow strip of the caudal fin) were taken from live individuals after fishes were anesthetized using MS-222 (Merck KGaA, Darmstadt, Germany).
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Responsible Editor: Lucia Carbone
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Štundlová, J., Hospodářská, M., Lukšíková, K. et al. Sex chromosome differentiation via changes in the Y chromosome repeat landscape in African annual killifishes Nothobranchius furzeri and N. kadleci. Chromosome Res 30, 309–333 (2022). https://doi.org/10.1007/s10577-022-09707-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-022-09707-3