Abstract
The presence or absence of Mad1 at kinetochores is a major determinant of spindle assembly checkpoint (SAC) activity, the surveillance mechanism that delays anaphase onset if one or more kinetochores remain unattached to spindle fibers. Among the factors regulating the levels of Mad1 at kinetochores is the Rod, Zw10, and Zwilch (RZZ) complex, which is required for Mad1 recruitment through a mechanism that remains unknown. The relative dynamics and interactions of Mad1 and RZZ at kinetochores have not been extensively investigated, although Mad1 has been reported to be stably recruited to unattached kinetochores. In this study, we directly compare Mad1-green fluorescent protein (GFP) turnover dynamics on unattached Drosophila kinetochores with that of RZZ, tagged either with GFP-Rod or GFP-Zw10. We find that nearly 40 % of kinetochore-bound Mad1 has a significant dynamic component, turning over with a half-life of 12 s. RZZ in contrast is essentially stable on unattached kinetochores. In addition, we report that a fraction of RZZ and Mad1 can co-immunoprecipitate, indicating that the genetically determined recruitment hierarchy (in which Mad1 depends on RZZ) may reflect a physical association of the two complexes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The spindle assembly checkpoint (SAC) is a surveillance system that delays anaphase onset if one or more kinetochores are not properly attached to the spindle (reviewed in Foley and Kapoor 2013; Hauf 2013; Jia et al. 2013; Lara-Gonzalez et al. 2012). This mechanism provides more time for the cell to correct the attachment and thus to avoid possible chromosome missegregation and consequent aneuploidy. Several outer kinetochore proteins, including Mad1, Mad2, BubR1, Bub3, Mps1, and the RZZ complex participate in the SAC and are necessary for generating the mitotic checkpoint complex, an assembly of Mad2, BubR1, Bub3 and the protein Cdc20, which is believed to be the anaphase inhibitor.
The presence or absence of Mad1 at kinetochores is a major determinant of SAC activity (Ballister et al. 2014; Maldonado and Kapoor 2011). Mad1, in complex with Mad2, can be viewed as a catalyst that helps generate the anaphase inhibitor (De Antoni et al. 2005; Fava et al. 2011; Sironi et al. 2001). Mad1/Mad2 levels are high on unattached kinetochores, but upon the establishment of proper kinetochore-microtubule (K-MT) attachment, Mad1 and Mad2 are removed in a dynein-dependent manner (Hoffman et al. 2001; Howell et al. 2000, 2001, 2004), in a process called variously stripping, streaming, or shedding. This dynein-mediated removal is believed to contribute to the extinction of the inhibitory signal generated by the SAC (Buffin et al. 2005; Howell et al. 2001; Wojcik et al. 2001). However, if Mad1 is artificially forced to remain on kinetochores even after attachment, its presence is sufficient to generate the anaphase inhibitor and maintain the cell in metaphase (Ballister et al. 2014; Maldonado and Kapoor 2011).
Despite Mad1’s critical role in the regulation of the SAC, the factors influencing its levels on kinetochores are only poorly understood. Mad1 appears to interact directly with Bub1 (London and Biggins 2014; Moyle et al. 2014), while the kinases Mps1 and Aurora B have also been implicated in Mad1 kinetochore targeting (Ditchfield et al. 2003; Lan and Cleveland 2010). In metazoan cells, Mad1/Mad2 recruitment to kinetochores additionally requires the activity of the Rod, Zw10, and Zwilch (RZZ) complex (Buffin et al. 2005; Karess 2005; Kops et al. 2005). All three subunits of RZZ are required for a functional SAC (Buffin et al. 2005; Scaerou et al. 2001; Williams et al. 2003). How RZZ might help recruit Mad1/Mad2 is unclear, as no direct or indirect physical association between Mad1 and RZZ has been reported to date.
Many of the kinetochore proteins involved in the SAC are highly dynamic on unattached kinetochores (Howell et al. 2000, 2004; Shah et al. 2004), displaying turnover times on the order of tens of seconds in the cases of Cdc20, Mps1, a portion of Mad2, and BubR1. However, Mad1 was in contrast reported to be mostly stable on unattached kinetochores (Howell et al. 2004; Shah et al. 2004). RZZ turnover has been less studied. In Drosophila, it was described as having a half-life of about 1 min on unattached kinetochores as measured in embryos (Basto et al. 2004), whereas in mammalian cells, RZZ’s association with kinetochores was reported to be essentially stable, at least after inhibition of microtubule polymerization in cells with a vinblastin treatment (Famulski et al. 2008).
Here, using fluorescently tagged Mad1 and Rod and the new reagent green fluorescent protein (GFP)-Zw10, we describe for the first time Mad1 turnover dynamics on Drosophila kinetochores and re-evaluate the dynamics of RZZ. We find that kinetochore-bound Mad1 has a significant dynamic component, representing nearly 40 % of the total, with a turnover time of 12 s, whereas RZZ is in fact stable on unattached kinetochores. In addition, we report that a fraction of RZZ and Mad1 can co-immunoprecipitate, indicating that the genetically determined dependence of Mad1 recruitment on RZZ can be linked to a physical association between the two complexes.
Methods
Transgenic flies
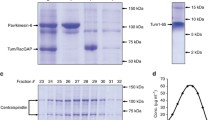
Transgenic Drosophila lines expressing GFP-zw10 were obtained by P-element transposition of a construct. The transgene corresponds to a 5-kb zw10 genomic sequence, including the promoter region, contained within two BamHI sites (Fig. 1) cloned into the vector CasPer4 (Thummel and Pirrotta 1992). The GFP tag is fused to the N-terminus of the Zw10 protein. Fly transformation was carried out by BestGene (Chino Hills, CA, USA). Spc25-GFP (described in Schittenhelm et al. 2007) was a gift from Christian Lehner (University of Zurich, Switzerland). RFP-Rab6 (Januschke et al. 2007), and ptc-GAL4 UAS-GFP (Legent et al. 2012) were gifts from Antoine Guichet and Kevin Legent, respectively (both at Institut Jacques Monod, Paris, France). GFP-Rod (Basto et al. 2004), RFP-Rod (Buffin et al. 2005), Mad1-GFP (Emre et al. 2011), and Cherry-Mad1 (Emre et al. 2011) have been previously described.
Schematic of the zw10 genomic region, and the transgene used in this study. A 5-kb BamHI genomic fragment spanning the zw10 promoter region and transcription unit was assembled into the CasPer4 transposon vector. Klp3A and bzd are genes adjacent to zw10 in the genome. The GFP tag was inserted just upstream of the ATG
Analysis of GFP-zw10 activity
For assaying cytokinesis failure during meiosis in spermatocytes, testes from young adult males of genotype zw10 5 or zw10 5; GFP-zw10 were dissected and mounted in 0.7 % saline, gently squashed between slide and coverslip and observed by phase-contrast microscopy for the presence of multinucleate spermatids (Williams et al. 1996). For counting abnormal anaphases, brains from zw10 5 or zw10 5; GFP-zw10 third instar larvae were fixed and stained in aceto-orcein and observed as described previously (Buffin et al. 2007). For assaying SAC function, brains were fixed and stained after incubation in colchicine (10−4 M) for 5 or 60 min, and the mitotic density (the average number of mitotic figures per microscope field with at least one mitosis) was determined (Buffin et al. 2007).
Antibodies
For western blots, anti-GFP mouse monoclonal (a mixture of clones 7.1 and 13.1, Roche Diagnostics, Meylan, France) was used at 1/1000; rabbit anti-Rod (Scaerou et al. 2001) was used at 1/6000; rabbit anti-Zw10 (Williams et al. 1992) was used at 1/1000; and mouse polyclonal anti-Mad1 (Emre et al. 2011) was used at 1/500. Secondary antibodies, horseradish peroxidase (HRP) anti-mouse (Sigma Chemical Co., Saint-Quentin Fallavier, France) and HRP anti-rabbit (Promega, Charbonnieres, France) were used as described by the suppliers. Western blots were performed as described in (Rubin et al. 2014).
Co-immunoprecipitation and proteomic analysis
All immunoprecipitation (IP) experiments were performed on protein extracts obtained from fresh embryos. One- to two-hour-old syncytial embryos expressing the GFP-tagged Rod or GFP-Mad1 transgenes were harvested, devitellinized, and treated with colchicine (100 μM) for 25 min at room temperature. Embryos expressing free GFP from a fly stock of genotype ptc-GAL4 UAS-GFP were used as a control. The embryos were lysed in homogenization buffer (10 mM Tris-HCl at pH 7.5, 0.15 M NaCl, 1 mM EDTA, 0.5 % NP40, complete ULTRA and phoStop inhibitors (both from Roche Diagnostics) using a Dounce homogenizer, and the lysates were precleared by centrifugation (2 × 10 min at 20,000×g)). For co-immunoprecipitation studies and for mass spectrometry analysis, 2 mg of precleared protein extract was immunoprecipitated with 50 μl of μMACS anti-GFP MicroBeads (Miltenyi Biotec, Bergisch Gladbach, Germany) for 30 min at 4 °C. The mixture was applied onto a μColumn (Miltenyi) and allowed to run through by gravity flow as described by the supplier. The immobilized beads were washed 4× with 200 μl of homogenization buffer without NP40 and 2× with 200 μl of 20 mM Tris-HCl at pH 7.5 buffer. Proteins were eluted from the beads with 50 μl of 0.5 M NH4OH. This procedure routinely immunoprecipitated 70–80 % of the total GFP-tagged proteins present in the lysate.
The isolated immune complexes were digested overnight at 37 °C with sequencing grade trypsin (12.5 μg/ml, Promega) in 20 μl of 25 mM NH4HCO3. Digests were analyzed by a LTQ Velos Orbitrap coupled to an Easy nano-LC Proxeon system (Thermo Fisher Scientific, Illkirch, France). Data were processed with Proteome Discoverer 1.4 software (Thermo Fisher) coupled to an in-house Mascot search server (version 2.3.02, Matrix Science, Boston, MA). False discovery rates for peptide identification were estimated by the Percolator algorithm (Matrix Science). A threshold of 0.01 was used to consider a peptide as identified.
In vivo observation of larval neuroblasts
Fluorescent time-lapse videos were acquired with an Olympus IX-70 inverted microscope, a focused Xenon lamp, and an OrcaER CCD camera (Hamamatsu Photonics K.K., Massy, France), piloted by the Cell-R hardware and software system (Olympus, Rungis, France). Acquisition times per frame were 100 ms for GFP-Zw10 and 150 ms for RFP-Rod, and images were obtained as stacks of five planes at 1-μm intervals taken every 15 s with a ×60 NA 1.4 oil objective. For Fig. 4, images of a single plane were recorded every 5 s.
FRAP
Neuroblasts from Drosophila larval brains were treated 15 min with colchicine at 10−4 M. Fluorescence recovery after photobleaching (FRAP) experiments were performed using a confocal microscopy system (Zeiss Axiovert with LSM780; Zeiss, Oberkochen, Germany) equipped with a Plan-Apo ×63 NA 1.4 oil objective (during experiments with Mad1-GFP, Spc25-GFP, and GFP-Rod) or a Plan-Apo ×40 NA 1.3 oil objective (during experiments with GFP-Zw10). Imaging was controlled by Zeiss confocal software Zen 2012. A circular area of 4 μm diameter (encompassing all the kinetochores), or 0.9 μm (containing just a few kinetochores), was bleached with the 488 or 514 nm lines of an Argon laser at 25 mW. Images were acquired with a temporal resolution of 1 s (for Mad1-GFP) or 5 s (for Spc25-GFP, GFP-Rod, and GFP-Zw10) with a GaAsP detector (490–650 nm) (Zeiss). Fluorescence intensity in a region of interest was quantified using ImageJ (Schneider et al. 2012). The exponential kinetics of fluorescence recovery after photobleaching were analyzed by nonlinear regression fitting using Origin software (OriginLab, Northampton, MA). The different fluorescent recovery kinetics and percent recovery of Mad1-GFP relative to GFP-Rod, GFP-Zw10, and Spc25-GFP were not influenced by the size of the bleached area, indicating that the low recovery of Rod and Zw10 reflects low-turnover rate and not inadvertant photobleaching of the pool of unbound protein.
Results
A functional GFP-Zw10 transgene
The dynamics of RZZ complex in vivo were previously studied in Drosophila only by following a fluorescent-tagged Rod subunit (Basto et al. 2004; Buffin et al. 2005). However, the functions of these three proteins overlap only partially. The Zw10 subunit is a conserved component of a second complex involved in vesicle trafficking in all eukaryotes (Hirose et al. 2004; Arasaki et al. 2007; Sun et al. 2007; Schmitt 2010; Civril et al. 2010; Menant and Karess 2010). It thus makes sense that in Drosophila spermatocytes, Zw10, but not Rod, has been shown to be required for proper cytokinesis (Wainman et al. 2012; Williams et al. 1996). Because of these divergent properties of Rod and Zw10, it is important to compare directly the behavior of both proteins in the same cells. In pursuit of this goal, we generated transgenic lines of GFP-tagged Zw10, in a genomic construct expressed from the gene’s endogenous promoter (see “Methods”). We show here that this transgene is fully functional.
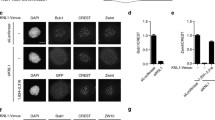
Two transgenic lines were obtained that partially rescued the zw10 5 null allele in one copy, and fully rescued when present in two copies. In interphase cells, GFP-Zw10 was found to be entirely cytoplasmic (Fig. 2), and to label vesicular structures that also express the Golgi marker RFP-Rab-6 (Januschke et al. 2007), similar to what has been described by immunostaining for Zw10 (Wainman et al. 2012). During mitosis, GFP-Zw10 labeled kinetochores and the spindle (see below). Dividing neuroblasts of rescued zw10 5 mutant flies carrying the GFP-Zw10 transgene showed none of the mitotic defects commonly seen in zw10 mutant animals: anaphases were normal and the cells displayed a normal SAC response (the average number of mitoses per optical field doubled after 1 h in colchicine (Table 1)). In addition, rescued flies showed no evidence of the spermatocyte cytokinesis failure found in zw10 mutants (Table 1) (Williams et al. 1996). By these criteria, the GFP-Zw10 fusion protein appears to substitute effectively for all functions previously ascribed to the endogenous Zw10 protein.
GFP-Zw10 localizes to Golgi vesicles in interphase cells. Two interphase spermatocytes are shown dissected from a male fly expressing GFP-Zw10 (green) and the Golgi marker RFP-Rab6 (red). GFP-Zw10 accumulates in a subset of the vesicles labeled by RFP-Rab6 (arrowheads, and merge, right). Scale bar, 5 μm
Co-recruitment and migration of Rod, Zw10, and Mad1
The GFP-Zw10 transgene allowed us to monitor independently the behaviors of Zw10 and Rod during mitosis. Figure 3 shows that GFP-Zw10 and RFP-Rod are simultaneously recruited to kinetochores at nuclear envelope breakdown (NEB). After alignment of chromosomes on the metaphase plate, both proteins are present in particles streaming from kinetochores, presumably along kinetochore-attached microtubules (K-MTs), (Fig. 3, insets). This finding confirms that the GFP-Zw10 protein is able to form a functional RZZ complex that recapitulates the activity of the endogenous protein complex during mitosis.
Simultaneous recruitment and streaming of RFP-Rod and GFP-Zw10. Frames from a movie of a zw10 5 ; GFP-zw10; RFP-rod larval neuroblast from just prior to NEB until anaphase. In interphase, both GFP-Zw10 and RFP-Rod are diffusely cytoplasmic, but Zw10 additionally is found in larger particles that we assume to be Golgi structures. Rod and Zw10 appear simultaneously on the kinetochores as the cell enters mitosis (at NEB). In metaphase, the two signals co-localize at the kinetochores as well as on particles migrating along the spindle fibers. The two proteins remain associated with kinetochores as they migrate to the poles during anaphase. Inset, detail of particles on kinetochore fibers, showing co-label of RFP-Rod and GFP-Zw10. Scale bars, 2 μm
We have previously reported that Mad2 and RZZ co-migrate in particles along K-MTs (Buffin et al. 2005). To determine whether Mad1 is also found in these migrating particles, we followed Mad1-GFP and RFP-Rod co-expressed in fly neuroblasts. Figure 4 (see also Movie 1) shows that indeed particles containing both Mad1 and RZZ can be seen co-migrating along K-MTs. As the streaming of outer kinetochore components has been shown to depend on dynein (Buffin et al. 2005; Howell et al. 2001; Wojcik et al. 2001), it is likely that these particles contain this microtubule motor as well as Mad1, Mad2, and RZZ.
Mad1 and RZZ stream together along kinetochore fibers. Frames from a movie of a metaphase neuroblast expressing RFP-Rod and Mad1-GFP in a wild-type background, taken on a single plane at 5-s intervals. Numerous small particles, visible in both channels, can be seen migrating from kinetochores to poles. The yellow arrowheads indicate a particularly prominent particle breaking off the kinetochore and migrating towards the pole. See also Movie 1 in Supplemental materials. Scale bar, 2 μm
A fraction of RZZ and Mad1 can co-immunoprecipitate
Mitotically active, syncytial stage Drosophila embryos (1–2-h-old) expressing Mad1-GFP in a mad1 null background were treated with colchicine and immunoprecipitated with anti-GFP. The anti-GFP antibody brought down not only Mad1-GFP but also a fraction of the Rod present in the extract (Fig. 5). Reciprocally, immunoprecipitates from embryos expressing GFP-Rod in a rod null background contained significant levels of Mad1. No Rod or Mad1 was detected in IPs of control embryo extracts (in which no transgene or only free GFP protein was expressed). Corroborating the western blot results shown in Fig. 5, mass spectrometry analysis (Table 2) of the immunoprecipitate of Mad1-GFP detected both Rod (mascot score 150) and Zw10 (mascot score 110), neither of which was detectable in the control IPs nor in interphase-enriched extracts.
Reciprocal co-immunoprecipitation of Rod and Mad1. GFP-tagged Rod (in a rod null mutant background) or Mad1 (in a mad1 null mutant background) expressed under their natural promoters in transgenic syncytial embryos, were immunoprecipitated by incubation with anti-GFP-specific antibody bound to magnetic beads, and the presence of co-immunoprecipitating proteins was revealed by specific antibodies on a western blot. Top left, Mad1-GFP immunoprecipitate brings down a portion of the available Rod. Bottom left, GFP-Rod immunoprecipitate brings down a portion of the available Mad1. Right columns, control anti-GFP immunoprecipitates of flies expressing free GFP or no GFP-tagged transgene at all do not bring down Rod or Mad1. The first lane of each pair is total lysate probed with anti-Rod or anti-Mad1 and corresponds to 1/50 of the extract volume used for the IP. Blots were exposed to generate total lysate signals of approximately equal intensities in the experimental and control extracts
The reciprocal co-IP of Rod and Mad1 indicates that there is likely to be a physical interaction (direct or indirect) between at least a subset of RZZ and Mad1 in mitotic cells. The relatively small fraction of RZZ and Mad1 that co-precipitates may also suggest that the interaction is either very weak, or highly transient.
FRAP analysis reveals different turnover dynamics of RZZ and Mad1 at kinetochores
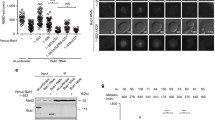
The FRAP technique allows determination of the half-life residency time of a fluorescent protein on a given structure. In order to examine the turnover dynamics of RZZ and Mad1 at unattached kinetochores, we performed FRAP experiments on Drosophila larval neuroblasts expressing either Mad1-GFP, GFP-Rod or GFP-Zw10 in a wild type background (Fig. 6).
FRAP analysis of kinetochore-bound RZZ and Mad1. Neuroblasts of Drosophila larval brains, treated 15 min with 10−4 M colchicine and expressing GFP-Rod, GFP-Zw10, Spc25-GFP, or Mad1-GFP were imaged before and after photobleaching of a small area covering one or two kinetochores (Rod and Zw10) or the entire kinetochore region (Spc25 and Mad1). Left, kinetics of fluorescence recovery. The recovery of signal in the bleached area was measured every 5 s and plotted as a fraction of the initial pre-bleach signal. Inset of Mad1, early-phase recovery of Mad1 measured every 1 s following bleach. Error bars are S.E.M. Right, examples of cells during photobleaching and recovery; the bleached areas are marked in yellow. Scale bars, 2 μm
Neither GFP-Rod nor GFP-Zw10 recovered more than 10 % of the initial pre-bleach signal levels, even after 8 min. This indicates that RZZ, once recruited, is stably associated with unattached kinetochores. In fact, its stability is essentially the same as that of Spc25-GFP (Schittenhelm et al. 2007), a subunit of the Ndc80 complex, which is known to be a stable component of M-phase kinetochores. We conclude that neither GFP-Rod nor GFP-Zw10 displays any appreciable turnover on unattached kinetochores.
In contrast to RZZ, photobleached kinetochore Mad1 rapidly recovered a significant fluorescent signal, reaching about 40 % of pre-bleach levels in about 30 s. The data fit well to a single exponential curve with a t ½ of about 12 s. The remainder of the Mad1 signal did not recover significantly during the duration of the analysis. Thus, two pools of Mad1 appear to exist at Drosophila kinetochores: one stable, and one rapidly turning over. This conclusion was unexpected because Mad1 had been reported previously to be a stable component of unattached mammalian kinetochores (Howell et al. 2004; Shah et al. 2004).
Discussion
We have shown here that at least some RZZ and Mad1 can be isolated in an immune complex from Drosophila, and that RZZ is stable on unattached kinetochores, while Mad1 is composed of two pools, one stable and one dynamic. We also examine GFP-Zw10 for the first time in Drosophila. GFP-Zw10 apparently retains the functionality of the endogenous protein both in Golgi and in mitosis. Moreover, it follows the same dynamics of GFP-Rod at the kinetochore, indicating that the kinetochore-associated RZZ complex is not disassembling during mitosis.
Two pools of Mad1, only one of RZZ
About 40 % of kinetochore-bound Mad1 displays a rapid turnover time more characteristic of the dynamic SAC components (Mad2, Cdc20, BubR1) than of the stable components (such as Bub1 and Bub3) (Howell et al. 2004). RZZ by contrast is almost entirely stable: in the absence of MTs, more than 90 % of kinetochore RZZ shows no recovery after 5 min, which is similar to the behavior of Spc25, a stable component of the outer kinetochore KMN (KNL-1, MIS12, NDC80) network. The fact that both Rod and Zw10 show the same low-to-nonexistent turnover argues that the RZZ complex remains intact at the kinetochore throughout mitosis. In an earlier study (Basto et al. 2004), we reported a moderate RZZ turnover. However, we now believe that conclusion to be erroneous, as the GFP-Rod signals were saturating the camera, and therefore not properly quantifiable. RZZ turnover dynamics on unattached kinetochores have not previously been investigated in other model systems, although mammalian RZZ was reported to be essentially stably associated with attached but tensionless kinetochores (treated with taxol) (Famulski et al. 2008).
Mad1 is usually considered to be a stable component of unattached mitotic kinetochores, based on two earlier FRAP analyses of Mad1 in PtK2 cells (Howell et al. 2004; Shah et al. 2004), both of which concluded that there is only a small dynamic pool. Howell et al. reported an average recovery of 29 ± 15 %; Shah et al. (2004) on the order of 10–12 %. However, careful examination of the graphs actually presented in Shah et al. suggests that the dynamic pool of Mad1 on unattached kinetochores might in fact be closer to the 40 % we find here. Moreover, a very recent study (Matson and Stukenberg 2014) using HeLa cells also reports that about 45 % of kinetochore Mad1 displays high turnover dynamics, and with a half life (18 s) similar to what we find in Drosophila (12 s). Thus, it would appear that a significant dynamic fraction of Mad1 is in fact a general feature of unattached kinetochores, and this fact needs to be considered in any model of kinetochore assembly and SAC function.
Why should there be two different populations, stable and dynamic, of kinetochore-bound Mad1? The two populations might differ by a post-translational modification such as phosphorylation. (Mammalian Mad1 is known to be phosphorylated at several residues during M phase (Bauer et al. 2014)). Possibly only one population (presumably the stable one (Maldonado and Kapoor 2011)) is competent to generate the SAC signal. Maybe Mad1 interacts with kinetochores in two steps: First it is recruited, but the connection remains unstable (an intermediate state), and subsequently the interaction is stabilized. For example, Bub1 which is known to bind to Mad1 on the outer kinetochore KMN complex (London and Biggins 2014; Moyle et al. 2014) might help stabilize Mad1 only after its initial recruitment. Whatever the explanation however, both the stable and dynamic pools of Mad1 ultimately depend on RZZ for their recruitment, since Mad1 kinetochore levels are near zero in the absence of RZZ.
In HeLa cells, RZZ recruitment to unattached kinetochores requires the Aurora B-dependent phosphorylation of the KMN component Zwint1 (Matson and Stukenberg 2014; Famulski and Chan 2007). The subsequent retention of RZZ and Mad1 at kinetochores appears to depend on the protein CenpI (Matson and Stukenberg 2014). However, neither Zwint1 nor CenpI seem to have Drosophila counterparts, or at least none has been identified by homology. It is therefore unclear whether these aspects of the regulation of recruitment and release of RZZ and Mad1 will be the same in flies.
Association of RZZ and Mad1
Although Mad1 recruitment to kinetochores depends on RZZ activity, the two complexes have not been previously reported to interact physically. Our successful co-immunoprecipitation of the two complexes from mitotic extracts is most likely biologically meaningful and not an artifact of lysis, because this association is mitosis-specific: Mad1-GFP immunoprecipitated from interphase-enriched Drosophila embryo extracts did not bring down any detectable Rod or Zw10 (Table 2).
Only a small fraction of the available RZZ and Mad1 co-precipitate, however. There may be several reasons for this. Firstly, the different turnover kinetics of Mad1 and RZZ at the kinetochore indicates that the two complexes are not necessarily in the same assembly. The interaction may involve only a subpopulation of Mad1, for example just the kinetochore-stable pool. Secondly, the interaction itself may be transient, or low-affinity, with only a small fraction of the low-turnover, kinetochore-bound Mad1 and RZZ interacting sufficiently stably to be isolated in an immune complex. Finally, not all of the Mad1 and Rod in a mitotic cell is kinetochore associated; some is found in the cytoplasm (see, for example, Fig. 4). If the interaction occurs only at kinetochores (a reasonable, but unproven assumption), it would not involve the cytoplasmic fractions.
The precise role of RZZ in recruiting Mad1 is not yet understood, but our results are consistent with a physical interaction (direct or indirect) between these two kinetochore components. It is worth noting that no other known kinetochore proteins were detected at significant levels by mass spectrometry, raising the possibility at least that there is no “missing link,” but just a direct low-affinity interaction.
Abbreviations
- SAC:
-
Spindle assembly checkpoint
- RZZ:
-
Rough deal-Zw10-Zwilch
- APC/C:
-
Anaphase promoting complex/cyclosome
- GFP:
-
Green fluorescent protein
- RFP:
-
Red fluorescent protein
- K-MT:
-
Kinetochore microtubule
- NEB:
-
Nuclear envelope breakdown
- IP:
-
Immunoprecipitation
- FRAP:
-
Fluorescence recovery after photobleaching
References
Arasaki K, Uemura T, Tani K, Tagaya M (2007) Correlation of Golgi localization of ZW10 and centrosomal accumulation of dynactin. Biochem Biophys Res Commun 359:811–816
Ballister ER, Riegman M, Lampson MA (2014) Recruitment of Mad1 to metaphase kinetochores is sufficient to reactivate the mitotic checkpoint. J Cell Biol 204:901–908
Basto R, Scaerou F, Mische S, Wojcik E, Lefebvre C, Gomes R, Hays T, Karess R (2004) In vivo dynamics of the rough deal checkpoint protein during Drosophila mitosis. Curr Biol 14:56–61
Bauer M, Ahrne E, Baron AP, Glatter T, Fava LL, Santamaria A, Nigg EA, Schmidt A (2014) Evaluation of data-dependent and -independent mass spectrometric workflows for sensitive quantification of proteins and phosphorylation sites. J Proteome Res 13:5973–5988
Buffin E, Lefebvre C, Huang J, Gagou ME, Karess RE (2005) Recruitment of Mad2 to the kinetochore requires the Rod/Zw10 complex. Curr Biol 15:856–861
Buffin E, Emre D, Karess RE (2007) Flies without a spindle checkpoint. Nat Cell Biol 9:565–572
Civril F, Wehenkel A, Giorgi FM, Santaguida S, Di Fonzo A, Grigorean G, Ciccarelli FD, Musacchio A (2010) Structural analysis of the RZZ complex reveals common ancestry with multisubunit vesicle tethering machinery. Structure 18:616–626
De Antoni A, Pearson CG, Cimini D, Canman JC, Sala V, Nezi L, Mapelli M, Sironi L, Faretta M, Salmon ED, Musacchio A (2005) The mad1/mad2 complex as a template for mad2 activation in the spindle assembly checkpoint. Curr Biol 15:214–225
Ditchfield C, Johnson VL, Tighe A, Ellston R, Haworth C, Johnson T, Mortlock A, Keen N, Taylor SS (2003) Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J Cell Biol 161:267–280
Emre D, Terracol R, Poncet A, Rahmani Z, Karess RE (2011) A mitotic role for Mad1 beyond the spindle checkpoint. J Cell Sci 124:1664–1671
Famulski JK, Chan GK (2007) Aurora B kinase-dependent recruitment of hZW10 and hROD to tensionless kinetochores. Curr Biol 17:2143–2149
Famulski JK, Vos L, Sun X, Chan G (2008) Stable hZW10 kinetochore residency, mediated by hZwint-1 interaction, is essential for the mitotic checkpoint. J Cell Biol 180:507–520
Fava LL, Kaulich M, Nigg EA, Santamaria A (2011) Probing the in vivo function of Mad1:C-Mad2 in the spindle assembly checkpoint. EMBO J 30:3322–3336
Foley EA, Kapoor TM (2013) Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat Rev Mol Cell Biol 14:25–37
Hauf S (2013) The spindle assembly checkpoint: progress and persistent puzzles. Biochem Soc Trans 41:1755–1760
Hirose H, Arasaki K, Dohmae N, Takio K, Hatsuzawa K, Nagahama M, Tani K, Yamamoto A, Tohyama M, Tagaya M (2004) Implication of ZW10 in membrane trafficking between the endoplasmic reticulum and Golgi. EMBO J 23:1267–1278
Hoffman DB, Pearson CG, Yen TJ, Howell BJ, Salmon ED (2001) Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol Biol Cell 12:1995–2009
Howell BJ, Hoffman DB, Fang G, Murray AW, Salmon ED (2000) Visualization of Mad2 dynamics at kinetochores, along spindle fibers, and at spindle poles in living cells. J Cell Biol 150:1233–1250
Howell BJ, McEwen BF, Canman JC, Hoffman DB, Farrar EM, Rieder CL, Salmon ED (2001) Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J Cell Biol 155:1159–1172
Howell BJ, Moree B, Farrar EM, Stewart S, Fang G, Salmon ED (2004) Spindle checkpoint protein dynamics at kinetochores in living cells. Curr Biol 14:953–964
Januschke J, Nicolas E, Compagnon J, Formstecher E, Goud B, Guichet A (2007) Rab6 and the secretory pathway affect oocyte polarity in Drosophila. Development 134:3419–3425
Jia L, Kim S, Yu H (2013) Tracking spindle checkpoint signals from kinetochores to APC/C. Trends Biochem Sci 38:302–311
Karess R (2005) Rod-Zw10-Zwilch: a key player in the spindle checkpoint. Trends Cell Biol 15:386–392
Kops GJ, Kim Y, Weaver BA, Mao Y, McLeod I, Yates JR 3rd, Tagaya M, Cleveland DW (2005) ZW10 links mitotic checkpoint signaling to the structural kinetochore. J Cell Biol 169:49–60
Lan W, Cleveland DW (2010) A chemical tool box defines mitotic and interphase roles for Mps1 kinase. J Cell Biol 190:21–24
Lara-Gonzalez P, Westhorpe FG, Taylor SS (2012) The spindle assembly checkpoint. Curr Biol 22:R966–R980
Legent K, Steinhauer J, Richard M, Treisman JE (2012) A screen for X-linked mutations affecting Drosophila photoreceptor differentiation identifies Casein kinase 1alpha as an essential negative regulator of wingless signaling. Genetics 190:601–616
London N, Biggins S (2014) Mad1 kinetochore recruitment by Mps1-mediated phosphorylation of Bub1 signals the spindle checkpoint. Genes Dev 28:140–152
Maldonado M, Kapoor TM (2011) Constitutive Mad1 targeting to kinetochores uncouples checkpoint signalling from chromosome biorientation. Nat Cell Biol 13:475–482
Matson DR, Stukenberg PT (2014) CENP-I and Aurora B act as a molecular switch that ties RZZ/Mad1 recruitment to kinetochore attachment status. J Cell Biol 205:541–554
Menant A, Karess R (2010) RZZ finds its ancestral roots. Structure 18:549–550
Moyle MW, Kim T, Hattersley N, Espeut J, Cheerambathur DK, Oegema K, Desai A (2014) A Bub1-Mad1 interaction targets the Mad1-Mad2 complex to unattached kinetochores to initiate the spindle checkpoint. J Cell Biol 204:647–657
Rubin T, Karess RE, Rahmani Z (2014) Cenp-meta is required for sustained spindle checkpoint. Biol Open 3:522–528
Scaerou F, Starr DA, Piano F, Papoulas O, Karess RE, Goldberg ML (2001) The ZW10 and Rough Deal checkpoint proteins function together in a large, evolutionarily conserved complex targeted to the kinetochore. J Cell Sci 114:3103–3114
Schittenhelm RB, Heeger S, Althoff F, Walter A, Heidmann S, Mechtler K, Lehner CF (2007) Spatial organization of a ubiquitous eukaryotic kinetochore protein network in Drosophila chromosomes. Chromosoma 116:385–402
Schmitt HD (2010) Dsl1p/Zw10: common mechanisms behind tethering vesicles and microtubules. Trends Cell Biol 20:257–268
Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675
Shah JV, Botvinick E, Bonday Z, Furnari F, Berns M, Cleveland DW (2004) Dynamics of centromere and kinetochore proteins; implications for checkpoint signaling and silencing. Curr Biol 14:942–952
Sironi L, Melixetian M, Faretta M, Prosperini E, Helin K, Musacchio A (2001) Mad2 binding to Mad1 and Cdc20, rather than oligomerization, is required for the spindle checkpoint. EMBO J 20:6371–6382
Sun Y, Shestakova A, Hunt L, Sehgal S, Lupashin V, Storrie B (2007) Rab6 regulates both ZW10/RINT-1 and conserved oligomeric Golgi complex-dependent Golgi trafficking and homeostasis. Mol Biol Cell 18:4129–4142
Thummel CS, Pirrotta V (1992) Technical notes: new pCasper P-element vectors. Drosophila Inf Serv 71:150
Wainman A, Giansanti MG, Goldberg ML, Gatti M (2012) The Drosophila RZZ complex—roles in membrane trafficking and cytokinesis. J Cell Sci 125:4014–4025
Williams B, Karr T, Montgomery J, Goldberg M (1992) The Drosophila l(1)zw10 gene product, required for accurate mitotic chromosome segregation is redistributed at anaphase onset. J Cell Biol 118:759–773
Williams B, Gatti M, Goldberg M (1996) Bipolar spindle attachments affect redistributions of ZW10, a Drosophila centromere/kinetochore component required for accurate chromosome segregation. J Cell Biol 134:127–140
Williams BC, Li Z, Liu S, Williams EV, Leung G, Yen TJ, Goldberg ML (2003) Zwilch, a new component of the ZW10/ROD complex required for kinetochore functions. Mol Biol Cell 14:1379–1391
Wojcik E, Basto R, Serr M, Scaerou F, Karess R, Hays T (2001) Kinetochore dynein: its dynamics and role in the transport of the Rough deal checkpoint protein. Nat Cell Biol 3:1001–1007
Acknowledgments
LD was supported by a fellowship from the “Ministère Français de l’enseignement supérieur et de la Recherche” and by fellowship ARC RK is Equipe Labellisée Ligue Contre le Cancer. Work at Cornell University (by BW and MG) was supported by NIH grant GM48430. We thank Antoine Guichet for RFP-Rab6, Christian Lehner for Spc25-RFP. We are very grateful to Thibault Leger and Camille Garcia of the Institut Jacques Monod Proteomics Platform. We also thank the ImagoSeine facility, a member of France BioImaging infrastructure, supported by the French National Research Agency (ANR-10-INSB-04, “Investments of the Future”).
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Conly Rieder
Electronic supplementary material
Below is the link to the electronic supplementary material.
Particles containing both RFP-Rod and Mad1-GFP stream from attached kinetochores (corresponding to Fig. 4; MOV 718 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made.
The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
To view a copy of this licence, visit https://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Défachelles, L., Raich, N., Terracol, R. et al. RZZ and Mad1 dynamics in Drosophila mitosis. Chromosome Res 23, 333–342 (2015). https://doi.org/10.1007/s10577-015-9472-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-015-9472-x