Abstract
Chromosome segregation during meiosis and mitosis depends on the assembly of functional kinetochores within centromeric regions. Centromeric DNA and kinetochore proteins show surprisingly little sequence conservation despite their fundamental biological role. However, our identification in Drosophila melanogaster of the most diverged orthologs identified so far, which encode components of a kinetochore protein network including the Ndc80 and Mis complexes, further emphasizes the notion of a shared eukaryotic kinetochore design. To determine its spatial organization, we have analyzed by quantitative light microscopy hundreds of native chromosomes from transgenic Drosophila strains coexpressing combinations of red and green fluorescent fusion proteins, fully capable of providing the essential wild-type functions. Thereby, Cenp-A/Cid, Cenp-C, Mis12 and the Ndc80 complex were mapped along the inter sister kinetochore axis with a resolution below 10 nm. The C terminus of Cenp-C was found to be near but well separated from the innermost component Cenp-A/Cid. The N terminus of Cenp-C is further out, clustered with Mis12 and the Spc25 end of the rod-like Ndc80 complex, which is known to bind to microtubules at its other more distal Ndc80/Nuf2 end.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The kinetochore which is formed within the centromeric region of eukaryotic chromosomes is crucial for faithful segregation of genetic information during mitotic and meiotic divisions. Its composition changes during the division cycle. In prometaphase, it allows attachment of chromosomes to spindle fibers (Rieder 2005). Moreover, it is associated with a number of checkpoint proteins that monitor chromosome integration into the spindle and prevent progression into anaphase as long as chromosomes without or with a syntelic attachment to the spindle are present.
Despite their fundamental biological role, centromeric DNA and primary sequences of associated proteins have evolved very rapidly (for recent reviews, see Schueler and Sullivan 2006; Vos et al. 2006). Initially, therefore, it has been difficult to integrate findings from different model organisms into a general model for kinetochore organization in eukaryotes. However, recent progress has dramatically improved the recognition of shared elements of centromere kinetochore complexes (CKC). The basis of CKC assembly appears to be formed by specialized chromatin containing nucleosomes with a centromere-specific histone H3 variant (Vos et al. 2006; Fujita et al. 2007). Cenp-A/Cid, the Drosophila centromere-specific histone H3 variant, is found at the centromere throughout the division cycles (Henikoff et al. 2000), as also true in other organisms (Vos et al. 2006). Cenp-C homologs represent another ubiquitous CKC component with a constitutive centromere localization (apart from the mitosis-specific association observed in the holocentric chromosomes of Caenorhabditis elegans; Moore and Roth 2001; Oegema et al. 2001; Heeger et al. 2005; Vos et al. 2006). Four multiprotein complexes (Sim4/COMA/NAC/Cenp-H/I, Mis12/MIND, Spc105/KNL-1, Ndc80) that were originally identified in yeast have also been characterized, at least partially, in a wide range of eukaryotes (McAinsh et al. 2003; Kline-Smith et al. 2005; Liu et al. 2005; Cheeseman et al. 2006; Foltz et al. 2006; Meraldi et al. 2006; Okada et al. 2006; Vos et al. 2006). Their centromere association in human cells appears to vary from constitutive (Sim4/COMA/NAC/Cenp-H/I; Foltz et al. 2006; McAinsh et al. 2006; Okada et al. 2006), constitutive except for telophase (Mis12; Kline et al. 2006; McAinsh et al. 2006), to mitosis-specific (Ndc80; Chen et al. 1997; Martin-Lluesma et al. 2002; Kline-Smith et al. 2005). Moreover, a number of mitotic spindle checkpoint proteins are conserved and present at high concentrations at those kinetochores, which are either not attached to the spindle or not exposed to the physical tension resulting from bipolar attachment (Vos et al. 2006). Apart from these proteins, about 50 additional CKC components have been described in various experimental systems, and the identification of diverged orthologs in other eukaryotes is progressing in many cases (Meraldi et al. 2006; Vos et al. 2006).
Initial insights into the details of the structural CKC organization were obtained by electron microscopy (EM; Rieder 1982; Vos et al. 2006). In Drosophila, kinetochore ultrastructure is similar to the appearance of vertebrate kinetochores (Maiato et al. 2006), in particular, during prometaphase when the hemispherical organization is transformed into a disk with a distinct outer plate of about 40 nm thickness. This outer plate is separated by a gap of about 30 nm from another electron-dense inner plate. In Drosophila, this inner plate is poorly resolved from the underlying amorphous inner chromatin mass. On the other side, i.e., distal to the outer plate, prometaphase kinetochores have a fibrous corona of variable depth (up to 200 nm), which has been clearly described in mammalian cells. We point out that kinetochore ultrastructure is known to change significantly from prophase to metaphase (Rieder 1982; Maiato et al. 2006), and that certain aspects like the distinction of the inner plate might reflect artifactual chromatin shrinking during fixation (McEwen et al. 1998).
Only a few CKC components have been localized by immuno-EM at an ultrastructural level. Thereby, vertebrate Cenp-C has been assigned to the inner plate (Saitoh et al. 1992) and the Ndc80 complex components Ndc80/Hec1 and Nuf2 to the outer plate (DeLuca et al. 2005). Moreover, Cenp-E has been located within the outer corona (Cooke et al. 1997). Based on double immunofluorescence comparisons with these well-studied proteins, almost all other CKC components have been tentatively classified as present either in the inner chromatin, the inner or outer plate, or the fibrous corona. Moreover, several studies have characterized kinetochore ultrastructure after RNA interference-mediated depletion of CKC components (Vigneron et al. 2004; DeLuca et al. 2005; Liu et al. 2006). RNA interference mostly in combination with fluorescence microscopy or immunodepletion in Xenopus egg extracts has also been used extensively to analyze dependencies in the CKC assembly process (see for instance Emanuele et al. 2005; Liu et al. 2006, and references in Vos et al. 2006). In general, inner components were found to be required for the later assembly of outer components, although certain inconsistencies point to a higher complexity (Liu et al. 2006). Finally, light microscopic analyses have been performed with stretched chromatin fibers, which have argued for a lateral association of repeating units into a kinetochore disk (Blower et al. 2002; Sullivan and Karpen 2004).
In this paper, we describe a light-microscopic approach for kinetochore analysis with unprecedented spatial resolution. This approach exploits some unique advantages of the model organism Drosophila melanogaster. Genetic complementation tests were used to demonstrate the functionality of CKC proteins fused to fluorescent proteins. These fluorescent proteins, the enhanced green fluorescent protein (EGFP) and monomeric red fluorescent protein (mRFP), provide localization tags, which are considerably smaller than antibodies. The cylindrical β-barrels formed by EGFP and mRFP have a diameter of 2.4 nm and a height of 4.2 nm (Ormo et al. 1996; Yang et al. 1996; Yarbrough et al. 2001). In contrast, indirect immunolocalization involves detection by two antibodies, each being more than fivefold larger than EGFP/mRFP, in combination with colloidal gold in the case of EM. In addition, fluorescent proteins eliminate potential problems with antibody specificity and antigen accessibility. Moreover, potential fixation artifacts can be avoided by imaging unfixed specimens. In Drosophila, the rapid syncytial division cycles of early embryogenesis result in a very high physiological mitotic index approaching 50%, which allows a very efficient preparation of native mitotic chromosomes. After our identification of components of the Drosophila Ndc80 and Mis12 complexes, we analyzed their localization in comparison to the previously described Cenp-A/Cid (Henikoff et al. 2000) and Cenp-C proteins (Heeger et al. 2005). By averaging the data obtained with hundreds of native chromosomes released from embryos coexpressing a red and a green fluorescent CKC component, we were able to map these proteins with a spatial resolution well beyond the light diffraction limit. Thereby, we localized Cenp-C to a region between the innermost Cenp-A/Cid and the outer Mis12 and Ndc80 complexes. Moreover, both Cenp-C and the rod-like Ndc80 complex were found to have a defined, polar orientation along the spindle axis.
Materials and methods
Fly strains
Drosophila stocks with the mutations Nuf2 SH2276 (Oh et al. 2003) and l(3)A34-1 (synonym: l(3)87Da 2; Hilliker et al. 1980) or the deficiencies Df(2L)ade3 and Df(3R)ry75 were obtained from the Bloomington stock center. The piggyBac insertion line Spc25 c00064 (Thibault et al. 2004) was kindly provided by the Harvard Medical School stock collection.
Nuf2 SH2276 appears to be a hypomorphic allele. Hemizygotes with Nuf2 SH2276 over Df(2L)ade3, which deletes Nuf2, were found to die during the late pupal stages, i.e., earlier than Nuf2 SH2276 homozygotes.
The EMS-induced recessive lethal mutation l(3)A34-1 which had genetically been mapped to a chromosomal region including Spc25 (Hilliker et al. 1980) failed to complement Spc25 c00064. Moreover, the lethality of l(3)A34-1 in trans over the deficiency Df(3R)ry75, which deletes Spc25, was rescued by the gSpc25-mRFP transgene. Spc25 c00064 homozygotes and hemizygotes (in trans over the deficiency Df(3R)ry75) displayed an indistinguishable phenotype which was more severe than that of l(3)A34-1 hemizygotes. Therefore, Spc25 c00064 appears to be an amorphic and l(3)A34-1 a hypomorphic allele.
The gene trap line P{w[+mC]=PTT-GA}Jupiter G00147 expresses a fusion of EGFP and Jupiter, a microtubule-binding protein (Morin et al. 2001). Transgenic strains expressing kinetochore proteins fused to fluorescent proteins were generated by standard P-element-mediated germ line transformation. Lines expressing functional Cenp-A/Cid with an internal EGFP insertion 11 amino acids before the start of the histone fold domain (gcid-EGFP-cid) have been described before (Schuh et al. 2007). Lines with an mRFP insertion in Cenp-A/Cid in place of the EGFP insertion (gcid-mRFP-cid) were generated and also confirmed to express a functional Cid fusion protein (S.H., unpublished observations). Lines expressing Cenp-C with an N-terminal EYFP extension (gEYFP-Cenp-C) or with a C-terminal EGFP extension (gCenp-C-EGFP) have been described before (Heeger et al. 2005). The gEGFP-Bub3, sryα-GAL4, and UAS-EGFP-CG11743 lines will be described in detail elsewhere. A line with sryα-GAL4 and UAS-EGFP-CG11743 was used for control in the coimmunoprecipitation experiments.
Additional lines were generated with the constructs described below. Lines with combinations of transgenes resulting in the expression of a red and a green fluorescent CKC component were generated by standard crosses. We analyzed lines with gcid-mRFP-cid II.1 in combination with either gcid-EGFP-cid III.2, gEYFP-Cenp-C II.1, gCenp-C-EGFP III.2, gEGFP-Nuf2 II.1, gMis12-EGFP II.1, gEGFP-Bub3, or P{w[+mC]=PTT–GA}Jupiter G00147. In addition, we also analyzed lines with gSpc25–mRFP II.1 in combination with either gSpc25-EGFP II.1, gcid-EGFP-cid III.2, gEYFP-Cenp-C II.1, gCenp-C-EGFP III.2, gEGFP-Nuf2 II.1, or gMis12-EGFP II.1. All transgenic lines had a w mutant background.
Plasmid constructions and transfections
pP{CaSpeR-4} constructs were made for the expression of kinetochore proteins fused to fluorescent proteins under control of the corresponding genomic regulatory region. Genomic fragments were amplified from bacterial artificial chromosome (BAC) clones characterized by the Drosophila genome project (BACR30C16 for Ndc80, BACR39J17 for Nuf2, BACR17F05 for Spc25, BACR14L10 for Mis12) (Hoskins et al. 2000). The sequences of the primers were: AnW20 (5′-G GAATTC GTA GAA TCG TTT GGA AAT GC-3′) and AnW21 (5′-G GGATCC CTT GGC GTT ATT GAA ACT AC-3′) for the 5′ part of Ndc80, AnW22 (5′-C TCTAGA ATG TCG CAC CTG ATG CCC-3′) and AnW23 (5′-CATTGT AGGCCT ACG TTA GCA CTA TCG GGG-3′) for the 3′ part of Ndc80; RaS83 (5′-CACCCAGTTC GCGGCCGC ATG TAT CAA ATG TGT CGC C-3′) and RaS84 (5′-GA GGATCC CAT TCA ATC CAG AGT TTT AAT-3′) for the 5′ part of Nuf2, RaS87 (5′-TG TCTAGA ATG GCG TTA TCA GTC GAA A-3′) and RaS88 (5′-A AGGCCT TGC CCC AGA TAA GGA AAA GG-3′) for the 3′ part of Nuf2; RaS93 (5′-GTTTAGATGG GCGGCCGC GCC GAT GAT CAG GAC CGG-3′) and RaS94 (5′-GG GGATCC GGT GTG GCT CAT CGG CG-3′) for the 5′ part of Spc25; RaS95 (5′-AC TCTAGA CTT CCG ATT AAC TGA TTT AC-3′) and RaS96 (5′-G AGGCCT CGA TTA ACA CCG GCC G-3′) for the 3′ part of Spc25; RaS125 (5′-AGC GAATTC GCT TCC TTT GTT TGT TCG GG-3′) and RaS126 (5′-GTT GGATCC ATC AGT CTC CTT CTT TAT CTG-3′) for the 5′ part of Mis12; RaS127 (5′-ACT TCTAGA ATA AAC TAA CTG GAT CAA GTT TT-3′) and RaS128 (5′-TCTCCCA AGGCCT CAG GCT TAT AGC AAA ATA TAC G-3′) for the 3′ part of Mis12. The polymerase chain reaction (PCR) fragments with the 5′ and 3′ parts of a given gene were introduced into polylinker sites of pP{CaSpeR-4}. Moreover, a PCR fragment containing the coding sequences of either mRFP1 or EGFP was introduced into the BamH1 and XbaI sites between the 5′ and 3′ parts. The primers for the amplification of the coding sequences of the fluorescent proteins were: RaS85 (5′-GC GGATCC ATG GTG AGC AAG GGC GAG-3′) and RaS86 (5′-TC TCTAGA CTT GTACAG CTC GTC CAT G-3′) for EGFP in gEGFP-Nuf2, RaS85 and RaS80 (5′-GG TCTAGA TTA CTT GTA CAG CTC GTC CAT G-3′) for EGFP in gMis12-EGFP and gSpc25-EGFP, RaS91 (5′-AG GGATCC ATG GCC TCC TCC GAG GAC-3′) and RaS92 (AA TCTAGA TTA GGC GCC GGT GGA GTG-3′) for mRFP1 in gSpc25-mRFP.
The pP{CaSpeR-4} constructs for expression of Cenp-C variants with green and red fluorescent proteins fused to N and C termini were based on a genomic 8.7 kb PmlI–XbaI fragment used previously for the generation of gEYFP-Cenp-C lines (Heeger et al. 2005). Inverse PCR with primers CM51 (5′-GTC GTT GCT AGC GGG CTT CGA CCT GAA AAA CAG-3′) and CM52 (5′-AAG CCC GCT AGC AAC GAC ACT CTG GAG CTG-3′) or MF8 (5′-GGC CTA GCT AGC ACT GCG TAT ACA CAT CAG CAC-3′) and MF9 (5′-CGC AGT GCT AGC TAG TAA TTG CTT TGT AAT TTA-3′) was used to introduce NheI sites after the start or directly before the stop codon, respectively. EGFP coding sequences were amplified with primers MF11 (5′-GGC CGC TAG CGT GAG CAA GGG CGA GGA GCT G-3′) and HS6 (5′-GGA CTA GTC TTG TAC AGC TCG TCC ATG C-3′) for N-terminal and MF11 and MF12 (5′-GGC CGC TAG CTT ACT TGT ACA GCT CGT CCA TG-3′) for C-terminal fusions. mRFP1 coding sequences were amplified with primers HSmRFP2 (5′-AGC GGC TAG CAT GGC CTC CTC CGA GGA C-3′) and HSmRFP3 (5′-CGA AAC TAG TGG CGC CGG TGG AGT GG-3′).
pPUAST constructs containing cDNAs fused to the EGFP coding sequence were used for transfection of Drosophila S2R+ cells. For these constructions, we first generated two pPUAST variants for either N- or C-terminal EGFP fusions. For the former variant, the EGFP coding sequence was amplified using the primers RaS42 (5′-C GAATTC ATG GTG AGC AAG GGC GAG-3′) and RaS43 (5′-TGGATTTCTG GCGGCCGC CTT GTA CAG CTC GTC CAT G-3′), which introduce an EcoRI and a NotI site before the initiation codon or the stop codon, respectively. This PCR fragment was introduced into the corresponding polylinker sites in pPUAST. For the latter variant, the EGFP coding sequence was amplified with the primers RaS79 (5′-GC GGTACC ATG GTG AGC AAG GGC GAG-3′) and RaS80 (5′-GG TCTAGA TTA CTT GTA CAG CTC GTC CAT G-3′), which introduce a KpnI and an XbaI site upstream of the initiation codon and after the stop codon, respectively. The resulting fragment was introduced into the corresponding polylinker sites of pPUAST. The restriction sites remaining in the polylinker of the pPUAST variants were used for the insertion of cDNA fragments amplified from expressed sequence tag (EST) plasmids characterized by the Drosophila genome project (Stapleton et al. 2002). The following EST plasmids were used: LD33040 (Ndc80), SD05495 (Nuf2), LD37196 (Spc25), RE19545 (Mis12), RE03006 (Kmn1), RE42502 (Nnf1a). Nnf1b and Kmn2 were amplified from genomic DNA. Primers used for amplification were: RaS74 (5′-GG GAATTC AT GCGGCCGC G ATG TCG CAC CTG ATG CC-3′) and RaS75 (5′-GG TCTAGA CTA ATG ATT CTT GAT GGC ATC TAG-3′) for pPUAST-EGFP-Ndc80, RaS77 (5′-GATTAAAACT GCGGCCGC A ATG GCG TTA TCA GTC GAA ATT-3′) and RaS78 (5′-TC TCTAGA TTA AGT GGA ATT CAT CTG CC-3′) for pPUAST-EGFP-Nuf2, RaS81 (5′-GG GGTACC GGT GTG GCT CAT CGG CG-3′) and RaS82 (5′-AG AGATCT ATG GCA ATT ATT ATG ACT GAA TC-3′) for pPUAST-Spc25-EGFP, RaS113 (5′-GTTA GCGGCCGC A ATG GAC TTC AAT AGC CTA GC-3′) and RaS114 (5′-AGTT GGTACC ATC AGT CTC CTT CTT TAT CTG-3′) for pPUAST-Mis12-EGFP, AnW28 (5′-ACGA GCGGCCGC T ATG GAG CCA GCC GAA AGT C-3′) and AnW30 (5′-GC GGTACC CCG TTG GTT GGC CAT ATT C-3′) for pPUAST-Kmn1-EGFP, AnW25 (5′-CAAA GCGGCCGC T ATG GAG GAT TCG GAA GCC G-3′) and AnW26 (5′-A CTCGAG TCA GAA GTC GTT CAA TGC-3′) for pPUAST-EGFP-Nnf1a, AnW36 (5′-TGTG GCGGCCGC A ATG AAT AAT ATT GAA GAG GAC AC-3′) and AnW37 (5′-TT GGTACC TTA CAT TTC TTC CTG CAC ATA C-3′) for pPUAST-EGFP-Nnf1b, RaS137 (5′-CAAAAA GCGGCCGC C ATG GAA AGT AAG CGC-3′) and RaS139 (5′-GA GGTACC CAG CAA GGA CAA GCA GTC C-3′) for pPUAST-Kmn2-EGFP.
Before transfection, S2R+ cells were re-plated in 24 well plates containing a coverslip. The culture medium (1 ml) was replaced after 12–24 h, and cells were transfected with a standard calcium phosphate precipitate containing 3 μg of plasmid DNA. Cells were fixed about 24–26 h after transfection and stained with antibodies against Cenp-A/Cid or Cenp-C and the DNA stain Hoechst 33258 essentially as described previously (Vass et al. 2003; Heeger et al. 2005).
Immunoprecipitation
For coimmunoprecipitation experiments, we collected 5–8 h embryos from either w*;P{w +, gEGFP-Nuf2} II.1 or w 1; P{w +, sryα-Gal4} II.1, P{w +, UAS-EGFP-CG11743} II.1 flies at 25°C. After dechorionization in 50% bleach, eggs were shock frozen in liquid nitrogen and stored at −80°C. 1 ml packed embryos were homogenized in 4 ml lysis buffer [50 mM 4-2-hydroxyethyl-1-piperazineethanesulfonic acid (HEPES) at pH 7.5, 60 mM NaCl, 3 mM MgCl2, 1 mM CaCl2, 0.2% Triton X-100, 0.2% Nonidet NP-40, 10% glycerol, 1 mM dithiothreitol (DTT)] containing 0.2 ml protease inhibitor cocktail (P 8340, Sigma). The extracts were cleared by centrifugation (20 min, 14000×g). For further clearing, supernatants were incubated with 0.15 ml Protein-A-Sepharose beads (Affi-Prep, Bio-Rad) during 1 h followed by another centrifugation. 25 μl Protein-A-Sepharose beads to which about 25 μg affinity-purified rabbit antibodies against GFP or mRFP (J. Dürr, S. Herzog and S. H., unpublished) had been crosslinked with dimethyl pimelimidate (Harlow and Lane 1988) were used for immunoprecipitation from the supernatant. Immunoprecipitates were washed four times with lysis buffer. During the third and fourth washes, 0.5% Nonidet NP-40 and 0.5% Triton-X-100 were present in the lysis buffer. Moreover, during the fourth wash, 300 mM NaCl was also present in the lysis buffer. Immunoprecipitated proteins were eluted for 5 min at 37°C with 0.04 ml 2% sodium dodecyl sulfate (SDS), 50 mM Tris–HCl, pH 7.5, followed by another elution at 94°C. Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer was added, and the immunoprecipitated proteins were resolved on a large 12% Tris–HCl precast gel (Bio-Rad). Silver staining was performed essentially as described (Blum et al. 1987). Fixation was done in 30% ethanol and 10% acetic acid. After a wash in 30% ethanol, water was used for an additional 20 min wash. 0.02 and 0.05% formaldehyde were included in the silver nitrate solution and the developer, respectively. Staining was terminated in 5% acetic acid. Excised bands were rinsed in water and analyzed by mass spectrometry essentially as described previously (Riedel et al. 2006). Briefly, bands were reduced with DTT, carboxymethylated using iodoacetamide, and digested with trypsin. Tryptic peptides were extracted with formic acid and separated by nano high-performance liquid chromatography (LC-Packings, Netherland) on a PepMap C 18 column. The eluate of the column was applied online to an LTQ ion trap mass spectrometer (Thermo-Fisher) equipped with a nanospray source. Mass data on all peptides and their fragmentation pattern were analyzed using the Mascot software (Matrix Science). Note that Fig. 2 displays proteins eluted at 37°C. The majority of EGFP-Nuf2 and Spc25-mRFP was only eluted during the second elution at 94°C.
Sequence comparison
Secondary structure predictions were performed using Quick2D (Biegert et al. 2006). In case of the human proteins, predictions are based on a multiple sequence alignment obtained with the position-specific iterated basic local alignment search tool (PSI-BLAST) search option. In case of the Drosophila proteins, predictions are based on a multiple sequence alignment assembled using ClustalW and ortholog sequences identified in Drosophilid genome sequences (Thompson et al. 1994; Richards et al. 2005; prepublication data from Agencourt Bioscience Corporation and Genome Sequencing Center at Washington University). In case of Drosophila Nnf1, the alignment contained both Nnf1a and Nnf1b paralog sequences, which result in essentially indistinguishable predictions when tested individually (data not shown). Nnf1 homologs in Drosophilid genomes were identified by BLAST searches and compared using ClustalW, which was also used for phylogenetic tree construction.
Preparation and analysis of native chromosomes
Eggs were collected on apple juice agar plates and aged to the syncytial blastoderm stages. After chorion removal with 50% bleach and extensive washing in water, eggs were returned to apple juice agar plates. Three eggs were lined up on a glass slide, and 3.5 μl of phosphate buffered saline containing 2 μg/ml Hoechst 33258 was added. The embryos were squashed by capillary forces after adding a coverslip (24 × 32 mm). Microscopic analyses were started within a few minutes after turbulent mixing within the specimen had settled. Single focal planes were acquired with a Zeiss Axioplan 2 imaging system using a 100× PlanApo NA 1.4 objective, an AxioCam MRm camera, and an AxioVision software. With this setup, a camera pixel represents about a 68 nm square region of the object. The extent of movement within the unfixed sample was controlled by comparing the spatial distribution of the DNA staining acquired before and after acquisition of the red and green signals. Chromosomes that had moved more than 0.2 μm during acquisition were excluded from the analyses. In addition, chromosomes which did not clearly display two distinct sister kinetochore signals were also excluded from the analyses. Thereby, chromosomes from anaphase embryos, as well as chromosomes from prometa- and metaphase embryos with an orientation of the axis between sister kinetochores perpendicular (or nearly perpendicular) to the slide were eliminated. The majority of chromosomes (about 70%, n = 436) from prometaphase and metaphase embryos displayed two distinct sister kinetochores. The squashing procedure therefore appears to result in a preferred planar orientation of the two sister kinetochore on the slide. The relatively weak signals in combination with the extent of bleaching observed especially with the mRFP fusion proteins prevented us from acquiring z stacks followed by three-dimensional analyses of centroid signal maxima. For evaluation, signal intensities along a line connecting the two sister kinetochores were determined using ImageJ software and transferred to MS Excel software for further analysis. The d RRinter, d GGinter and d RGintra values for a given pair of CKC components (see Fig. 7d and Table 1), as well as the d inter values obtained for a given CKC component by pooling the corresponding data from the pairwise analyses (Table 2), were not normally distributed, as expected since chromosomes with closely spaced, overlapping sister kinetochore signals were excluded from the analyses for instance. An adequate statistical error analysis was further complicated by unexplored potential biological (kinetochore variation in chromosome 2, 3, 4, X and Y, or precise mitotic stage), experimental (chromosome orientation and extent of stretch/compression), and instrumental variability (pixelation, background noise). The distribution of all the measured data is therefore given in Fig. 7f–i.
Results
Identification of Drosophila Ndc80 and Mis12/MIND complexes
Bioinformatic analysis revealed similarities between known Ndc80/Hec1 proteins and the predicted Drosophila CG9938 product (data not shown). Similar findings from a systematic bioinformatic search for eukaryotic kinetochore proteins were described while this work was in progress (Meraldi et al. 2006). However, the observed similarities are only very limited and not correlated with evolutionary descent. Vertebrate Ndc80/Hec1 proteins are clearly more similar to fungal and plant homologs than to CG9938. To confirm therefore that the diverged CG9938 indeed encodes the Drosophila Ndc80 homolog, we expressed an EGFP fusion protein in Drosophila S2R+ cells. The subcellular localization of the EGFP signals (data not shown) was found to correspond to the known behavior of human Ndc80/Hec1 (Chen et al. 1997; Martin-Lluesma et al. 2002). Moreover, the expected localization was subsequently also observed in transgenic embryos expressing EGFP-CG9938 fusion protein (Fig. 1a). During interphase, we did not observe signals above background (Fig. 1a). However, during mitosis, distinct signals were present at the kinetochore from prometaphase until late anaphase. The kinetochore signals were in close proximity to those obtained after double labeling with antibodies against the constitutive centromere protein Cenp-C (Fig. 1a). Based on this and the following evidence, we conclude that CG9938 represents the Drosophila Ndc80 gene.
Localization of Drosophila Ndc80 complex components and Mis12. Transgenic embryos expressing EGFP fused to either a Ndc80, b Nuf2, c Spc25, or d Mis12 were fixed at the stage when some cells progress through the 14th, asynchronous round of mitosis. Cells in interphase (left panels) and metaphase (right panels) after double labeling with an antibody against Cenp-C (Cenp-C) and a DNA stain (DNA) illustrate that the EGFP fusion proteins were only associated with centromeres during mitosis. Bar = 5 μm
In a genome-wide yeast two hybrid screen, Drosophila Ndc80 was reported to support a high confidence interaction with the predicted CG8902 product (Giot et al. 2003). Moreover, this CG8902 protein has a very limited similarity to the Ndc80 complex component Nuf2 (Meraldi et al. 2006). To evaluate whether this most distant Nuf2 family member displayed the expected kinetochore localization during mitosis, it was also expressed as an EGFP fusion protein in S2R+ cells. Its subcellular localization (data not shown) was found to be indistinguishable from that of vertebrate Ndc80 components. In addition, the same localization behavior was also observed in transgenic embryos expressing EGFP-CG8902 (Fig. 1b). We conclude that CG8902 represents the Drosophila Nuf2 gene.
An additional Drosophila Ndc80 interactor (Giot et al. 2003), the CG7242 product, was observed to have limited similarity to the Ndc80 complex component Spc25. This gene was also identified as the most distant Spc25 family member in the bioinformatic study (Meraldi et al. 2006). An analysis of the intracellular localization of an EGFP fusion protein confirmed the expected kinetochore localization also in this case, both in transfected S2R+ cells (data not shown) and in transgenic embryos (Fig. 1c). We conclude that CG7242 encodes Drosophila Spc25.
Apart from Ndc80 complex members, the bioinformatic search had revealed an additional putative Drosophila CKC component. The CG18156 protein was reported to have very limited sequence similarity to fungal and metazoan Mis12 proteins (Meraldi et al. 2006). Mis12 was originally identified in fission yeast and subsequently found to be a component of a conserved CKC complex called Mis12/MIND, which contains three additional proteins (Takahashi et al. 1994; De Wulf et al. 2003; Goshima et al. 2003; Obuse et al. 2004; Kline et al. 2006). Our localization studies after transfection of S2R+ cells with an EGFP-CG18156 expression construct revealed centromere localization. EGFP signals were found to be colocalized with Cenp-A/Cid at the centromere not only during mitosis but also in interphase cells (Fig. 3a). This localization behavior corresponds to that of human Mis12, which is centromeric throughout the cell cycle except for a brief period in late telophase and early G1 (Kline et al. 2006; McAinsh et al. 2006). In transgenic embryos expressing EGFP-CG18156, we observed the centromeric signals only during mitosis (Fig. 1d). The apparent discrepancy concerning localization during interphase in transfected cells and transgenic embryos might reflect differences in expression levels and background signals. Primary and secondary structure comparisons (Fig. 4a) and kinetochore localization strongly support the proposal that CG18156 encodes the Drosophila Mis12 homolog.
To demonstrate that the identified putative Drosophila Ndc80 complex components Ndc80, Nuf2, and Spc25 are indeed present in a complex in vivo, we used mass spectrometry to analyze the proteins coprecipitated with functional EGFP-Nuf2 or Spc25-mRFP from embryo extracts. Antibodies against EGFP and mRFP were used for immunoprecipitation. SDS-PAGE of the immunoprecipitates followed by silver staining revealed the presence of proteins specifically coprecipitated by EGFP-Nuf2 but not by an unrelated control EGFP fusion protein (Fig. 2a,b). Mass spectrometry revealed the identity of these proteins. Among the coimmunoprecipitated proteins were Drosophila Ndc80 and Spc25, as expected from the Ndc80 complex characterizations in yeast and vertebrates (Kline-Smith et al. 2005). Moreover, Drosophila Mis12 was also coimmunoprecipitated. In addition, we detected the products of the uncharacterized CG13434 and CG1558 in the EGFP-Nuf2 immunoprecipitates (Fig. 2a). An expression of corresponding EGFP fusion protein in S2R+ cells revealed a kinetochore localization in both cases (Fig. 3b,d). CG13434 appears to encode a Drosophila homolog of Nnf1, which is a Mis12 complex component in yeast, vertebrates, and C. elegans (De Wulf et al. 2003; Nekrasov et al. 2003; Westermann et al. 2003; Cheeseman et al. 2004; Obuse et al. 2004; Kline et al. 2006). While primary structure comparisons revealed only very limited similarity (data not shown), secondary structure comparisons (Fig. 4a) provided additional support for our suggestion that CG13434 encodes a Drosophila Nnf1 homolog. Interestingly, the sequenced genomes of melanogaster subgroup species (D. melanogaster, D. simulans, D. sechellia, D. erecta, D. yakuba) all encode a CG13434 paralog, resulting from an apparent duplication of the primordial CG13434 ortholog at the start of the melanogaster subgroup lineage (Fig. 4b). An EGFP fusion of this paralog, CG31658, was also observed to localize to the centromere during interphase and mitosis (Fig. 3c). Therefore, we will designate CG13434 as Nnf1a and CG31658 as Nnf1b. Reverse transcriptase (RT)-PCR experiments suggested coexpression of the Nnf1 paralogs at least during early Drosophila development (Fig. 4c). The coimmunoprecipitation experiments with Spc25-mRFP confirmed an association of the Ndc80 complex components (Ndc80, Nuf2, Spc25) with Mis12, Nnf1a, and Nnf1b (Fig. 2b; data not shown).
Coimmunoprecipitation of Ndc80 and MIND complex components. Antibodies against EGFP or mRFP were used for immunoprecipitation from extracts of embryos expressing either EGFP-Nuf2 (Nuf2), Spc25-mRFP (Spc25) or an unrelated EGFP fusion protein (Con) for control. After SDS-PAGE and silver staining, selected bands were characterized by mass spectrometry. Numbers at margins indicate the molecular weight (kDa) of proteins in the marker lane (MW). a Bands enriched in the EGFP-Nuf2 immunoprecipitates (in comparison to control immunoprecipitates) were found to contain either only highly abundant cellular proteins (mostly ribosomal proteins) presumably reflecting nonspecific contaminations (stars) or also some rare proteins (as measured by their representation in EST data bases; arrowheads). As indicated by the names on the left margin, these rare proteins were found to be either components of the Ndc80 complex or putative Mis/MIND complex components. b In search of a potential Spc24 homolog, the molecular weight range from 10–35 kDa with proteins in Spc25-mRFP immunoprecipitates was fractioned in slices and analyzed. The position and names of rare proteins (as measured by their representation in EST data bases) which were subsequently confirmed to be kinetochore proteins (see text) are indicated
Localization of additional Drosophila kinetochore network components in Schneider cells. S2R+ cells were transfected with constructs allowing expression of EGFP fused to either Drosophila a Mis12, b Nnf1a, c Nnf1b, d Kmn1, and e Kmn2. Fixed cells in interphase (left panels) and metaphase (right panels) after double labeling with an antibody against Cenp-A/Cid (Cenp-A) and a DNA stain (DNA) illustrate that association of EGFP fusion proteins with centromeres is either observed during a–d interphase and mitosis or e restricted to mitosis. Bar = 5 μm; no bar visible
Secondary structure and genomic comparison of Drosophila kinetochore proteins. a Secondary structure predictions reveal similarities between Drosophila (Dm) and human (Hs) homologs of Mis12, Nnf1, and between Drosophila Kmn2 and the C-terminal region of human Spc24. Each dot represents an amino acid position. The lines with colored regions illustrate predictions obtained with various algorithms (PSIPRED, JNET, PROF Quali and King, PROF Rost, COILS from top to bottom). α-helical regions are shown in red, regions with β-sheets in blue and coiled coils in magenta. b A phylogenetic tree was constructed after aligning the predicted amino acids sequences encoded by Nnf1-like genes in Drosophilid genomes. An Nnf1 gene duplication resulting in the two paralogs Nnf1a and Nnf1b early after the divergence of the melanogaster subgroup lineage provides the most parsimonious explanation for the observed branching pattern. D. melanogaster (Dmel), D. simulans (Dsim), D. sechellia (Dsec), D. yakuba (Dyak), D. erecta (Dere), D. ananassae (Dana), D. persimilis (Dper), D. pseudoobscura (Dpse), D. willistoni (Dwil). Synteny considerations (data not shown) indicate that Nnf1a represents the primordial homolog. c The developmental expression pattern of D. melanogaster Nnf1a and Nnf1b was analyzed by RT-PCR experiments. The stages analyzed were: embryos 0–2 (0–2), 2–4 (2–4), 4–8 (4–8), and 8–16 (8–16) hours after egg deposition; larval stages (L1, L2, L3), pupae (P), adult males (M) and females (F). Control amplifications (G) from a cloned Nnf1a cDNA and an intron containing genomic Nnf1b fragment, as well as amplifications (N) with mock reverse transcribed mRNA demonstrated that the RT-PCR products were not derived from contaminating genomic DNA. The results are consistent with Nnf1a expression being correlated with mitotic proliferation and Nnf1b expression being germline-specific
The CG1558 product, which was coimmunoprecipitated with both EGFP-Nuf2 and Spc25-mRFP (Fig. 2), did not display significant similarities to known kinetochore network proteins in its predicted primary and secondary structure. After expression as an EGFP fusion protein, it was detected at the centromeres in both interphase and mitotic S2R+ cells (Fig. 3d). This localization behavior was identical to Mis12 but different from Ndc80, which displayed a mitosis-specific kinetochore localization. Therefore, CG1558 might encode an additional Mis12 complex subunit. Based on its size, it might correspond to Nsl1. However, both Nsl1 and Dsn1 are very poorly conserved Mis12 complex components according to a comparison of human, C. elegans, and yeast sequences. CG1558 will be designated as Kmn1 (kinetochore Mis12-Ndc80 network component 1).
Spc24 is present as a fourth 22–24 kDa subunit in both yeast and vertebrate Ndc80 complexes. We did not identify a potential Drosophila Spc24 homolog in the EGFP-Nuf2 and Spc25-mRFP immunoprecipitates within the corresponding molecular weight range. However, we detected (Fig. 2b) an 11 kDa product of the predicted gene HDC12388 (Hild et al. 2003), which is not annotated in FlyBase. Expression of an EGFP fusion protein in S2R+ cells revealed kinetochore localization during mitosis (Fig. 3e). The C. elegans Ndc80 complex is thought to contain a rudimentary 11 kDa Spc24 version (Cheeseman et al. 2006, supporting speculations that HDC12388 might correspond to Drosophila Spc24. According to secondary structure predictions, the HDC12388 product might correspond to the C-terminal globular Spc24 domain (Fig. 4a). At present, HDC12388 will be designated as Kmn2.
More extensive purification of the Drosophila Ndc80 and Mis12 complexes will be required for their complete characterization. However, our findings demonstrate that Drosophila expresses a network of interacting kinetochore proteins including particularly diverged homologs of the Mis12 and Ndc80 complexes. Although hardly evident from simple sequence comparisons, Drosophila kinetochore organization obviously shares extensive similarities with yeast, C. elegans, and vertebrates, which all have a kinetochore protein network containing the Mis12 and Ndc80 complexes (Wigge and Kilmartin 2001; De Wulf et al. 2003; Goshima et al. 2003; McCleland et al. 2003; Nekrasov et al. 2003; Westermann et al. 2003; Cheeseman et al. 2004; Obuse et al. 2004; Emanuele et al. 2005; Liu et al. 2005; Kline et al. 2006).
Essential functions of Drosophila Ndc80 complex components can be provided by fluorescent fusion protein variants
To define the functions of the Drosophila Ndc80 complex genetically, we identified and characterized mutations in Nuf2 and Spc25 (Fig. 5). A P element insertion in Nuf2 had been isolated in a transposon mutagenesis screen for recessive lethal mutations (Oh et al. 2003). The insertion SH2276 was confirmed to be located 11 bp upstream of the initiation codon in the 5′ untranslated region (UTR) of Nuf2. The insertion leads to a partial loss of function. Nuf2 SH2276 homozygotes were found to develop to the late pupal stages and to die either as pharate adults or soon after eclosion. The gEGFP-Nuf2 transgene was found to prevent the lethality of early Nuf2 SH2276 adults. Our findings demonstrate that Drosophila Nuf2 is an essential gene. Moreover, they indicate that the EGFP-Nuf2 fusion protein is functional.
Genetic characterization of Ndc80 and putative MIND complex components. Gene models are shown for a Ndc80/CG9938, b Nuf2/CG8902, c Spc25/CG7242, and d Mis12/CG18156. Moreover, transposon insertions present in mutant alleles are indicated by triangles above the gene models. In addition, the genomic fragments used for transgene constructions are indicated by the solid lines below the gene models. The position where the EGFP or mRFP coding sequence was inserted in these transgenes is illustrated by the triangles below the gene models
The piggyBac transposon insertion c00064 (Thibault et al. 2004) was mapped 38 bp upstream of the initiation codon in the 5′ UTR of Spc25. Spc25 c00064 homozygotes were found to die during the late larval stages. Compared to sibling control larvae, the mutants had only rudimentary imaginal discs and small brains, suggesting that Spc25 is required in mitotically proliferating cells. The late larval lethality of Spc25 c00064 homozygotes was prevented by a transgene driving expression of a Spc25-mRFP fusion protein under control of the Spc25 regulatory region (gSpc25-mRFP). This transgene also complemented the recessive lethality of Spc25 A34−1, another independent allele previously isolated as a recessive lethal mutation (Hilliker et al. 1980). Based on these observations, we conclude that Drosophila Spc25 is an essential gene. In addition, the Spc25-mRFP fusion protein must be functional.
Mapping of centromere and kinetochore proteins in native chromosomes at high resolution
The accuracy of distance measurements can be increased beyond the diffraction-limited resolution of light microscopy by analyzing the spatial separation between signals from two different fluorophores (Stelzer 1998; Shimogawa et al. 2006). Therefore, we constructed a number of strains that coexpressed both a red and a green fluorescent CKC component. Eggs were collected from these strains and aged to the syncytial blastoderm stage where thousands of nuclei progress synchronously through mitoses. Embryos were gently squashed in a buffer containing a DNA stain. Native chromosomes released from mitotic embryos were analyzed by wide-field fluorescence microscopy. Analyses with embryos expressing red fluorescent Cenp-A/Cid and a green fluorescent microtubule binding protein demonstrated that spindles did not survive the squashing procedure and that sister kinetochores of the released chromosomes were not under tension (Fig. 6a).
Association of released native chromosomes with spindles and mitotic checkpoint proteins. a To evaluate whether and how kinetochores of released native chromosomes interact with spindle remnants, we performed experiments with embryos expressing a green fluorescent microtubule binding protein (G147) and red fluorescent Cenp-A/Cid (Cenp-A). 82.4% of the chromosomes (n = 319) had kinetochores which showed no association with microtubules, as illustrated by the inset in the upper left corner which displays the kinetochores indicated by the filled arrowhead at high magnification. 16.6% had kinetochores with a lateral association with microtubules, as illustrated by the inset in the lower right corner which displays the kinetochores indicated by the open arrowhead. 0.3% had kinetochores with a monopolar end-on attachment. 0.6% appeared to have a bipolar attachment with microtubules which however appeared to be so disorganized that the attachment presumably did not result in significant tension across the sister kinetochores. Bar = 5 μm. b Native chromosomes were released from embryos expressing red fluorescent Cenp-A/Cid and green fluorescent Bub3. In contrast to Cenp-A/Cid, the localization of EGFP-Bub3 in kinetochores from different chromosomes was found to be highly variable, as illustrated by the kinetochores indicated by numbered arrowheads in the top panels and shown at high magnification in the lower panels. Bar = 5 μm
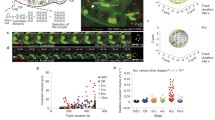
Pixel intensities of red and green CKC signals in native chromosomes released from prometa- or metaphase embryos were quantified along the axis connecting the two sister kinetochores (Fig. 7a–c). The distance between the intensity maxima in the two sister kinetochores was determined. The difference between the distances separating the green (d GGinter) and red (d RRinter) fluorescent maxima, respectively, was calculated and divided by two to obtain an estimate for the distance (d RGintra) between the red and the green fluorescent component within a CKC (Fig. 7d). Independent estimates for d RGintra from at least 100 different chromosomes released from at least five different embryos were averaged to yield an estimate of the distance which separates a given pair of CKC proteins (Table 1). These pair separation values are represented by double arrows in Fig. 7e. Moreover, as an additional estimate for the spatial arrangement of the CKC proteins, we determined the average of all d GGinter and d RRinter values obtained for a given CKC protein in all our analyses. For instance, in the case of Cenp-A/Cid, the 500 d RRinter measurements and 200 d GGinter measurements resulted in an average value for the separation of the two sister kinetochore Cenp-A/Cid signal maxima of 460 nm. From the separation differences apparent when different CKC components were compared (Table 2), the CKC components were positioned along the centromere-spindle axis as indicated by the vertical colored lines in Fig. 7e.
Axial positions of different kinetochore proteins along the inter sister kinetochore axis. a Native chromosomes released from syncytial embryos coexpressing a red and a green fluorescent CKC component were labeled with a DNA stain (DNA). The appearance of the native chromosomes with red (Cenp-Ar) and green (Cenp-Ag) fluorescent Cenp-A/Cid is illustrated at low magnification. Bar = 5 μm. b The fourth chromosome indicated by the white arrows in a is shown at high magnification. Bar = 0.5 μm. c A fourth chromosome from an embryo expressing red fluorescent Cenp-A/Cid (Cenp-Ar) and green fluorescent Nuf2 (Nuf2g) is shown at the same magnification as in b. These high magnification views, b and c, display the pixel resolution as acquired. d For an estimation of the spatial separation between a red and a green fluorescent CKC component, signal intensities were quantified along the axis running through the two sister kinetochores, as illustrated by the white line in c. The displayed intensity profiles are from the chromosome shown in c. The spatial separation (d RGintra) was calculated by halving the difference between the distances separating the red (d RRinter) and green (d GGinter) signal maxima of the sister kinetochores, as indicated by the equation. e Scheme summarizing the positions of the analyzed CKC components along the spindle axis. Double arrows represent the spatial separation as revealed by the pairwise analyses outlined in a–d with hundreds of native chromosomes (see also Table 1). Arrows above the upper dashed line represent data from experiments comparing the distance between red fluorescent Cenp-A/Cid and various green fluorescent CKC components. Analogous analyses of the distance between various green fluorescent CKC components and red fluorescent Spc25 are represented by the arrows between the dashed lines. The arrows below the lower dashed lines represent experiments after expression of Cenp-C versions labeled with both a green and a red fluorescent protein at the N and C termini. Color coding specifies the green fluorescent component in all these pairwise analyses. The vertical lines indicate the position of CKC components as revealed by pooling all d RRinter and d GGinter values obtained for a given CKC component during the pairwise analyses (see Table 2). The numbers indicate the spatial separation (nm) from the innermost centromere component Cenp-A/Cid, which was set to zero. In case of Cenp-C, the N terminus (N) and the C terminus (C) were mapped. f–i Histogram curves illustrate the distribution of the d RGintra values obtained in the pairwise analyses with chromosomes from embryos expressing red fluorescent Cenp-A/Cid and green CKC components, in f, as well as in the analogous analyses with red fluorescent Spc25 in combination with green CKC components, in g, or with Cenp-C versions carrying red and green fluorescent proteins at N and C termini, in h. Moreover, i the distribution of the d inter values, i.e., all the d RRinter and d GGinter measurements obtained for a given CKC components (see Table 2) are displayed as well
To determine the position of a spindle checkpoint protein in native chromosomes, we used a strain coexpressing red fluorescent Cenp-A/Cid and green fluorescent Bub3. Various mitotic spindle checkpoint components have been reported to localize to the outermost, fibrous corona region of the kinetochore in intact cells (Vos et al. 2006). In our native chromosome preparations, the localization of EGFP-Bub3 was found to be highly variable on different kinetochores (Fig. 6b), precluding a precise mapping.
The following control experiments support the accuracy of our CKC map. We generated a strain in which one of the two transgenes generated green and the other red fluorescent Cenp-A/Cid. In native chromosomes prepared from this strain (Fig. 7a,b), the red or green fluorescent proteins are expected to be perfectly colocalized. Our distance measurement resulted in d RGintra = 5 nm (Table 1). In an analogous analysis with a strain coexpressing red and green fluorescent Spc25, we obtained the expected d RGintra = 0 nm (Table 1). These distances were far smaller than the d RGintra values obtained for pairs of different proteins (Table 1). Moreover, the separation between signals from fluorescent protein tags at the N and the C termini of Cenp-C was determined twice using embryos expressing either EGFP-Cenp-C-mRFP or the reverse tag configuration mRFP-Cenp-C-EGFP, yielding 15 and 8 nm, respectively (Table 1). Finally, the additive behavior of different d RGintra values was found to correspond closely to the expectations. For instance, the sum of the measured distances between Cenp-A/Cid and Spc25 signals (d RGinter = 49 nm) on one hand and Spc25 and Nuf2 signals (d RGinter = 15 nm) on the other hand is only 5 nm different from the measured separation between Cenp-A/Cid and Nuf2 signals (d RGinter = 69 nm). Based on all these findings, we expect the correct signal maxima to be within ±5 nm of the positions indicated in Fig. 7e. We emphasize that our CKC map (Fig. 7e) was obtained by averaging values with distributions illustrated in Fig. 7f–i. As discussed below, averaging has important consequences for the interpretation of our CKC map.
Discussion
Our identification of Drosophila kinetochore proteins further exposes hidden similarities of kinetochore design in eukaryotes. In addition to the previously known, highly diverged Cenp-A/Cid and Cenp-C homologs (Henikoff et al. 2000; Heeger et al. 2005), Drosophila expresses similarly diverged homologs of the Mis12 and Ndc80 complex network, which is also present in yeast, C. elegans, vertebrates, and presumably in plants as well (Sato et al. 2005; Meraldi et al. 2006; Vos et al. 2006). We have been able to position several of these ubiquitous CKC components along the intersister kinetochore axis with unprecedented spatial resolution. Early Drosophila embryos allow an efficient isolation of native mitotic chromosomes and thereby imaging with reduced background. Moreover, transgenic strains allow the expression of fluorescent fusion proteins, which were demonstrated to be fully functional by genetic complementation tests.
We have determined the position of fluorescent signal maxima within the kinetochore of native chromosomes released from embryos expressing fluorescent CKC fusion proteins. Our CKC map (Fig. 7e) is based on averaged data from hundreds of analyzed chromosomes. Therefore, its interpretation depends critically on the variability of kinetochore organization in individual chromosomes. For instance, in principle, a given component might be localized on the inner kinetochore side in 50% of the chromatids and on the outer side in the other half of the chromatids, resulting in a misleading central positioning in our CKC map. Theoretically, such variability should widen the distribution of the distances measured in individual chromosomes (Fig. 7f–i). However, kinetochore width is smaller than the spreading of the image of a point light source in the microscope, and several additional factors (like background, noise, pixelation) further limit the precision of our measurements. The effect of positional variability on distribution width of the measured values would therefore be very subtle. Moreover, none of the known CKC proteins has been firmly demonstrated to be a spatially invariable kinetochore component, precluding comparisons to an established standard distribution. However, the reproducible trilaminar structure of the kinetochore during prometaphase which has been documented by EM, argues strongly against extensive organizational variability. We emphasize that the difficulties to detect subtle alterations in the distribution width of the measurements obtained for a given CKC component has important consequences even under the assumption that the spatial distribution of CKC components is essentially invariable in individual kinetochores. These difficulties prevent conclusions concerning the width occupied by a given CKC component within a kinetochore. For instance, Mis12 could either be confined to a single layer in the middle of the kinetochore or spread throughout the kinetochore, and both localization patterns would result in a central signal maximum. However, biochemical analyses of kinetochore proteins, which will be discussed in part below, have so far revealed highly specific interactions, arguing strongly for a precise and restricted localization of CKC components. The following discussion is therefore based on the unproven but likely assumption that the kinetochore represents a precisely defined layered structure.
Based on previous analyses, Cenp-A, Cenp-C, and Mis12 are thought to be components of the inner plate of the characteristic trilaminar kinetochore structure apparent in the EM (Kline et al. 2006; Vos et al. 2006). Our analyses indicate a significant separation between the inner most CKC component Cenp-A and all other CKC components analyzed here. Recently, Cenp-A nucleosomes purified from human cells were found to be intimately associated with the five proteins Cenp-M, Cenp-N, Cenp-T, Cenp-U, and Cenp-H in addition to Cenp-C (Foltz et al. 2006; Izuta et al. 2006; Okada et al. 2006). The apparent space between Cenp-A and Cenp-C might therefore be occupied by some of those proteins.
Many immunolocalization studies, including a recent study with Drosophila cells (Maiato et al. 2006), have failed to detect a comparable extensive spatial separation between Cenp-A/Cid and Cenp-C. However, immunolocalization with human chromosomes also revealed little overlap between Cenp-A and Cenp-C, with the latter extending over the top and bottom of a Cenp-A cylinder (Blower et al. 2002). Antigen accessibility problems, which were not excluded by Blower et al. (2002), cannot affect our concurrent findings.
In this paper, Cenp-C is shown to be spread in a polar orientation across a central CKC region. The C-terminal domain of Cenp-C, which contains the most conserved region including the CENP-C motif (Talbert et al. 2004; Heeger et al. 2005), points toward the centromeric DNA. These C-terminal sequences are connected via minimally conserved spacer sequences to the N-terminal domain which is oriented toward the kinetochore spindle fibers. The N-terminal region of D. melanogaster Cenp-C contains some blocks which are highly conserved among Drosophilids (Heeger et al. 2005). These blocks might be involved in recruiting the next layer of kinetochore proteins which we suggest to include the Ndc80 and Mis12 complexes. We find Mis12 to be close to the N-terminal Cenp-C region. Moreover, the Ndc80 complex component Spc25 appears to be even a bit closer but well separated by about 20 nm from the other Ndc80 component Nuf2. Apart from a polar Cenp-C orientation, our analyses therefore also indicate a polar orientation for the Ndc80 complex.
The tetrameric Ndc80 complex has a highly elongated, rod-like structure in vitro (Ciferri et al. 2005; Wei et al. 2005). The globular N-terminal domains of Ndc80 and Nuf2 are present on one end of the rod. The remainder of these two subunits forms an extended coiled coil which is further prolonged at its C-terminal end by binding to the N-terminal coiled coil region of the Spc24/Spc25 dimer. Closely associated C-terminal globular domains of Spc24 and Spc25 (Wei et al. 2006) form the other end of the rod. Scanning force microscopy and EM analyses have indicated that the coiled coil region separating the globular domains at the end of the Ndc80 complex has an extension of about 40 nm (Ciferri et al. 2005; Wei et al. 2005). This is twofold longer than the distance that we have observed between fluorescent proteins at the N and C termini of Nuf2 and Spc25 in kinetochores of native Drosophila chromosomes. Many of the elongated Ndc80 complexes might not be perfectly oriented along the spindle axis, especially as the kinetochores in our preparations are not under tension. Such a nonuniform orientation could result in spatial distributions of the N and C termini of Nuf2 and Spc25, respectively, with signal maxima that are more closely spaced than their separation within an isolated complex. An analysis of the positions of CKC components in chromosomes that are bi-oriented within the spindle and under tension would clearly be of interest. However, the increased background levels present in living embryos have so far precluded such analyses.
Our observed polar orientation of the Ndc80 complex within the kinetochore confirms the findings of a recent independent study (Deluca et al. 2006). Moreover, the observation that Ndc80 and Nuf2 kinetochore localization is no longer observed in the absence of Spc24 or Spc25 (Bharadwaj et al. 2004) is consistent but does not prove an orientation of the complex with inner Spc24/Spc25 and outer Ndc80/Nuf2 globular domains, because absence of Spc24 or Spc25 for instance might simply result in an instability of other complex components, as often observed in the case of stable complexes.
In budding yeast, the Ndc80 complex has been proposed to function as a connection between the inner components (CBF3 complex, Cenp-A/Cse4 nucleosome, Cenp-C/Mif2, Mis12/MIND complex) and the Dam/DASH complex which is required for bi-orientation and appears to form a ring around the single microtubule attaching to a yeast kinetochore (Cheeseman et al. 2001; Shang et al. 2003; Tanaka et al. 2005; Westermann et al. 2006). More recently, bacterial expression of the C. elegans KMN network composed of the Spc105/KNL-1, Mis12 and Ndc80 complexes has led to a convincing identification of two independent sites in this protein network which can bind directly to microtubules in vitro (Cheeseman et al. 2006). One of these microtubule binding sites is present within Spc105/KNL-1. The other is found within the globular N-terminal Ndc80 domain (Cheeseman et al. 2006) which is known to be within the outer kinetochore plates where kinetochore microtubules terminate (DeLuca et al. 2005). In vitro, the Ndc80 complex binds to microtubules at an angle (Cheeseman et al. 2006). A corresponding orientation of the Ndc80 complex within the kinetochore is fully consistent with our finding that the separation of the terminal globular domains of Spc25 and Nuf2 along the intersister kinetochore axis appears to be less than their separation along the axis of isolated complexes (Ciferri et al. 2005; Wei et al. 2005). Accordingly, the “barbed end” of microtubules decorated with the Ndc80 complex would be predicted to correspond to the plus end.
In conclusion, in addition to the identification of Drosophila Ndc80 and Mis12 complex components, our work provides a highly resolved structural framework integrating the most widely studied ubiquitous CKC components and a precise method for a future incorporation of additional proteins.
References
Bharadwaj R, Qi W, Yu H (2004) Identification of two novel components of the human NDC80 kinetochore complex. J Biol Chem 279:13076–13085
Biegert A, Mayer C, Remmert M, Soding J, Lupas AN (2006) The MPI bioinformatics toolkit for protein sequence analysis. Nucleic Acids Res 34:W335–W339
Blower MD, Sullivan BA, Karpen GH (2002) Conserved organization of centromeric chromatin in flies and humans. Dev Cell 2:319–330
Blum H, Beier H, Gross HJ (1987) Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 8:93–99
Cheeseman IM, Brew C, Wolyniak M, Desai A, Anderson S, Muster N, Yates JR, Huffaker TC, Drubin DG, Barnes G (2001) Implication of a novel multiprotein Dam1p complex in outer kinetochore function. J Cell Biol 155:1137–1145
Cheeseman IM, Niessen S, Anderson S, Hyndman F, Yates JR 3rd, Oegema K, Desai A (2004) A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev 18:2255–2268
Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A (2006) The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell 127:983–997
Chen Y, Riley DJ, Chen PL, Lee WH (1997) HEC, a novel nuclear protein rich in leucine heptad repeats specifically involved in mitosis. Mol Cell Biol 17:6049–6056
Ciferri C, De Luca J, Monzani S, Ferrari KJ, Ristic D, Wyman C, Stark H, Kilmartin J, Salmon ED, Musacchio A (2005) Architecture of the human ndc80–hec1 complex, a critical constituent of the outer kinetochore. J Biol Chem 280:29088–29095
Cooke CA, Schaar B, Yen TJ, Earnshaw WC (1997) Localization of CENP-E in the fibrous corona and outer plate of mammalian kinetochores from prometaphase through anaphase. Chromosoma 106:446–455
DeLuca JG, Dong Y, Hergert P, Strauss J, Hickey JM, Salmon ED, McEwen BF (2005) Hec1 and nuf2 are core components of the kinetochore outer plate essential for organizing microtubule attachment sites. Mol Biol Cell 16:519–531
Deluca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, Salmon ED (2006) Kinetochore microtubule dynamics and attachment stability are regulated by hec1. Cell 127:969–982
De Wulf P, McAinsh AD, Sorger PK (2003) Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes Dev 17:2902–2921
Emanuele MJ, McCleland ML, Satinover DL, Stukenberg PT (2005) Measuring the stoichiometry and physical interactions between components elucidates the architecture of the vertebrate kinetochore. Mol Biol Cell 16:4882–4892
Foltz DR, Jansen LE, Black BE, Bailey AO, Yates JR 3rd, Cleveland DW (2006) The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol 8:458–469
Fujita Y, Hayashi T, Kiyomitsu T, Toyoda Y, Kokubu A, Obuse C, Yanagida M (2007) Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev Cell 12:17–30
Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, Vijayadamodar G, Pochart P, Machineni H, Welsh M, Kong Y, Zerhusen B, Malcolm R, Varrone Z, Collis A, Minto M, Burgess S, McDaniel L, Stimpson E, Spriggs F, Williams J, Neurath K, Ioime N, Agee M, Voss E, Furtak K, Renzulli R, Aanensen N, Carrolla S, Bickelhaupt E, Lazovatsky Y, DaSilva A, Zhong J, Stanyon CA, Finley RL Jr, White KP, Braverman M, Jarvie T, Gold S, Leach M, Knight J, Shimkets RA, McKenna MP, Chant J, Rothberg JM (2003) A protein interaction map of Drosophila melanogaster. Science 302:1727–1736
Goshima G, Kiyomitsu T, Yoda K, Yanagida M (2003) Human centromere chromatin protein hMis12, essential for equal segregation, is independent of CENP-A loading pathway. J Cell Biol 160:25–39
Harlow E, Lane D (1988) Antibodies. A laboratory manual. Cold Spring Harbor Laboratory, New York
Heeger S, Leismann O, Schittenhelm R, Schraidt O, Heidmann S, Lehner CF (2005) Genetic interactions of Separase regulatory subunits reveal the diverged Drosophila Cenp-C homolog. Genes Dev 19:2041–2053
Henikoff S, Ahmad K, Platero JS, van Steensel B (2000) Heterochromatic deposition of centromeric histone H3-like proteins. Proc Natl Acad Sci USA 97:716–721
Hild M, Beckmann B, Haas SA, Koch B, Solovyev V, Busold C, Fellenberg K, Boutros M, Vingron M, Sauer F, Hoheisel JD, Paro R (2003) An integrated gene annotation and transcriptional profiling approach towards the full gene content of the Drosophila genome. Genome Biol 5:R3
Hilliker AJ, Clark SH, Chovnick A, Gelbart WM (1980) Cytogenetic analysis of the chromosomal region immediately adjacent to the rosy locus in Drosophila melanogaster. Genetics 95:95–110
Hoskins RA, Nelson CR, Berman BP, Laverty TR, George RA, Ciesiolka L, Naeemuddin M, Arenson AD, Durbin J, David RG, Tabor PE, Bailey MR, DeShazo DR, Catanese J, Mammoser A, Osoegawa K, de Jong PJ, Celniker SE, Gibbs RA, Rubin GM, Scherer SE (2000) A BAC-based physical map of the major autosomes of Drosophila melanogaster. Science 287:2271–2274
Izuta H, Ikeno M, Suzuki N, Tomonaga T, Nozaki N, Obuse C, Kisu Y, Goshima N, Nomura F, Nomura N, Yoda K (2006) Comprehensive analysis of the ICEN (interphase centromere complex) components enriched in the CENP-A chromatin of human cells. Genes Cells 11:673–684
Kline-Smith SL, Sandall S, Desai A (2005) Kinetochore-spindle microtubule interactions during mitosis. Curr Opin Cell Biol 17:35–46
Kline SL, Cheeseman IM, Hori T, Fukagawa T, Desai A (2006) The human Mis12 complex is required for kinetochore assembly and proper chromosome segregation. J Cell Biol 173:9–17
Liu X, McLeod I, Anderson S, Yates JR 3rd, He X (2005) Molecular analysis of kinetochore architecture in fission yeast. EMBO J 24:2919–2930
Liu ST, Rattner JB, Jablonski SA, Yen TJ (2006) Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J Cell Biol 175:41–53
Maiato H, Hergert PJ, Moutinho-Pereira S, Dong Y, Vandenbeldt KJ, Rieder CL, McEwen BF (2006) The ultrastructure of the kinetochore and kinetochore fiber in Drosophila somatic cells. Chromosoma 115:469–480
Martin-Lluesma S, Stucke VM, Nigg EA (2002) Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science 297:2267–2270
McAinsh AD, Tytell JD, Sorger PK (2003) Structure, function, and regulation of budding yeast kinetochores. Annu Rev Cell Dev Biol 19:519–539
McAinsh AD, Meraldi P, Draviam VM, Toso A, Sorger PK (2006) The human kinetochore proteins Nnf1R and Mcm21R are required for accurate chromosome segregation. EMBO J 25:4033–4049
McCleland ML, Gardner RD, Kallio MJ, Daum JR, Gorbsky GJ, Burke DJ, Stukenberg PT (2003) The highly conserved Ndc80 complex is required for kinetochore assembly, chromosome congression, and spindle checkpoint activity. Genes Dev 17:101–114
McEwen BF, Hsieh CE, Mattheyses AL, Rieder CL (1998) A new look at kinetochore structure in vertebrate somatic cells using high-pressure freezing and freeze substitution. Chromosoma 107:366–375
Meraldi P, McAinsh AD, Rheinbay E, Sorger PK (2006) Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol 7:R23
Moore LL, Roth MB (2001) HCP-4, a CENP-C-like protein in Caenorhabditis elegans, is required for resolution of sister centromeres. J Cell Biol 153:1199–1208
Morin X, Daneman R, Zavortink M, Chia W (2001) A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proc Natl Acad Sci USA 98:15050–15055
Nekrasov VS, Smith MA, Peak-Chew S, Kilmartin JV (2003) Interactions between centromere complexes in Saccharomyces cerevisiae. Mol Biol Cell 14:4931–4946
Obuse C, Iwasaki O, Kiyomitsu T, Goshima G, Toyoda Y, Yanagida M (2004) A conserved Mis12 centromere complex is linked to heterochromatic HP1 and outer kinetochore protein Zwint-1. Nat Cell Biol 6:1135–1141
Oegema K, Desai A, Rybina S, Kirkham M, Hyman AA (2001) Functional analysis of kinetochore assembly in Caenorhabditis elegans. J Cell Biol 153:1209–1226
Oh SW, Kingsley T, Shin HH, Zheng Z, Chen HW, Chen X, Wang H, Ruan P, Moody M, Hou SX (2003) A P-element insertion screen identified mutations in 455 novel essential genes in Drosophila. Genetics 163:195–201
Okada M, Cheeseman IM, Hori T, Okawa K, McLeod IX, Yates JR 3rd, Desai A, Fukagawa T (2006) The CENP-H–I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat Cell Biol 8:446–457
Ormo M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ (1996) Crystal structure of the Aequorea victoria green fluorescent protein. Science 273:1392–1395
Richards S, Liu Y, Bettencourt BR, Hradecky P, Letovsky S, Nielsen R, Thornton K, Hubisz MJ, Chen R, Meisel RP, Couronne O, Hua S, Smith MA, Zhang P, Liu J, Bussemaker HJ, van Batenburg MF, Howells SL, Scherer SE, Sodergren E, Matthews BB, Crosby MA, Schroeder AJ, Ortiz-Barrientos D, Rives CM, Metzker ML, Muzny DM, Scott G, Steffen D, Wheeler DA, Worley KC, Havlak P, Durbin KJ, Egan A, Gill R, Hume J, Morgan MB, Miner G, Hamilton C, Huang Y, Waldron L, Verduzco D, Clerc-Blankenburg KP, Dubchak I, Noor MA, Anderson W, White KP, Clark AG, Schaeffer SW, Gelbart W, Weinstock GM, Gibbs RA (2005) Comparative genome sequencing of Drosophila pseudoobscura: chromosomal, gene, and cis-element evolution. Genome Res 15:1–18
Riedel CG, Katis VL, Katou Y, Mori S, Itoh T, Helmhart W, Galova M, Petronczki M, Gregan J, Cetin B, Mudrak I, Ogris E, Mechtler K, Pelletier L, Buchholz F, Shirahige K, Nasmyth K (2006) Protein phosphatase 2A protects centromeric sister chromatid cohesion during meiosis I. Nature 441:53–61
Rieder CL (1982) The formation, structure, and composition of the mammalian kinetochore and kinetochore fiber. Int Rev Cytol 79:1–58
Rieder CL (2005) Kinetochore fiber formation in animal somatic cells: dueling mechanisms come to a draw. Chromosoma 114:310–318
Saitoh H, Tomkiel J, Cooke CA, Ratrie H 3rd, Maurer M, Rothfield NF, Earnshaw WC (1992) CENP-C, an autoantigen in scleroderma, is a component of the human inner kinetochore plate. Cell 70:115–125
Sato H, Shibata F, Murata M (2005) Characterization of a Mis12 homologue in Arabidopsis thaliana. Chromosome Res 13:827–834
Schueler MG, Sullivan BA (2006) Structural and functional dynamics of human centromeric chromatin. Annu Rev Genomics Hum Genet 7:301–313
Schuh M, Lehner CF, Heidmann S (2007) Incorporation of Drosophila Cid/Cenp-A and Cenp-C into centromeres during early embryonic anaphase. Curr Biol 17:237–243
Shang C, Hazbun TR, Cheeseman IM, Aranda J, Fields S, Drubin DG, Barnes G (2003) Kinetochore protein interactions and their regulation by the Aurora kinase Ipl1p. Mol Biol Cell 14:3342–3355
Shimogawa MM, Graczyk B, Gardner MK, Francis SE, White EA, Ess M, Molk JN, Ruse C, Niessen S, Yates JR 3rd, Muller EG, Bloom K, Odde DJ, Davis TN (2006) Mps1 phosphorylation of Dam1 couples kinetochores to microtubule plus ends at metaphase. Curr Biol 16:1489–1501
Stapleton M, Carlson J, Brokstein P, Yu C, Champe M, George R, Guarin H, Kronmiller B, Pacleb J, Park S, Wan K, Rubin GM, Celniker SE (2002) A Drosophila full-length cDNA resource. Genome Biol 3:RESEARCH0080.0081-0080.0088
Stelzer EH (1998) Contrast, resolution, pixelation, dynamic range and signal-to-noise ratio: fundamental limits to resolution in fluorescence light microscopy. J Microsc 189:15–24
Sullivan BA, Karpen GH (2004) Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat Struct Mol Biol 11:1076–1083
Takahashi K, Yamada H, Yanagida M (1994) Fission yeast minichromosome loss mutants mis cause lethal aneuploidy and replication abnormality. Mol Biol Cell 5:1145–1158
Talbert PB, Bryson TD, Henikoff S (2004) Adaptive evolution of centromere proteins in plants and animals. J Biol 3:18
Tanaka K, Mukae N, Dewar H, van Breugel M, James EK, Prescott AR, Antony C, Tanaka TU (2005) Molecular mechanisms of kinetochore capture by spindle microtubules. Nature 434:987–994
Thibault ST, Singer MA, Miyazaki WY, Milash B, Dompe NA, Singh CM, Buchholz R, Demsky M, Fawcett R, Francis-Lang HL, Ryner L, Cheung LM, Chong A, Erickson C, Fisher WW, Greer K, Hartouni SR, Howie E, Jakkula L, Joo D, Killpack K, Laufer A, Mazzotta J, Smith RD, Stevens LM, Stuber C, Tan LR, Ventura R, Woo A, Zakrajsek I, Zhao L, Chen F, Swimmer C, Kopczynski C, Duyk G, Winberg ML, Margolis J (2004) A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat Genet 36:283–287
Thompson JD, Higgins DG, Gibson TJ (1994) ClustalW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Vass S, Cotterill S, Valdeolmillos AM, Barbero JL, Lin E, Warren WD, Heck MM (2003) Depletion of Drad21/Scc1 in Drosophila cells leads to instability of the cohesin complex and disruption of mitotic progression. Curr Biol 13:208–218
Vigneron S, Prieto S, Bernis C, Labbe JC, Castro A, Lorca T (2004) Kinetochore localization of spindle checkpoint proteins: who controls whom? Mol Biol Cell 15:4584–4596
Vos LJ, Famulski JK, Chan GK (2006) How to build a centromere: from centromeric and pericentromeric chromatin to kinetochore assembly. Biochem Cell Biol 84:619–639
Wei RR, Sorger PK, Harrison SC (2005) Molecular organization of the Ndc80 complex, an essential kinetochore component. Proc Natl Acad Sci USA 102:5363–5367
Wei RR, Schnell JR, Larsen NA, Sorger PK, Chou JJ, Harrison SC (2006) Structure of a central component of the yeast kinetochore: the Spc24p/Spc25p globular domain. Structure 14:1003–1009
Westermann S, Cheeseman IM, Anderson S, Yates JR 3rd, Drubin DG, Barnes G (2003) Architecture of the budding yeast kinetochore reveals a conserved molecular core. J Cell Biol 163:215–222
Westermann S, Wang HW, Avila-Sakar A, Drubin DG, Nogales E, Barnes G (2006) The Dam1 kinetochore ring complex moves processively on depolymerizing microtubule ends. Nature 440:565–569
Wigge PA, Kilmartin JV (2001) The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J Cell Biol 152:349–360
Yang F, Moss LG, Phillips GN Jr (1996) The molecular structure of green fluorescent protein. Nat Biotechnol 14:1246–1251
Yarbrough D, Wachter RM, Kallio K, Matz MV, Remington SJ (2001) Refined crystal structure of DsRed, a red fluorescent protein from coral, at 2.0-A resolution. Proc Natl Acad Sci USA 98:462–467
Acknowledgement
We thank Brigitte Jaunich for technical help, as well as Julia Dürr and Sabine Herzog for the help with antibody production. Research in the laboratory of Karl Mechtler was supported by the 6th Framework Programme of the European Union via the Integrated Project MitoCheck and the Austrian Proteomics Platform (APP) within the Austrian Genome Program (GENAU), Vienna, Austria. In addition, this work was also supported by the Fonds der Deutschen Chemie and by a grant from the Deutsche Forschungsgemeinschaft (DFG Le 987/3–3 and 3–4).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by E.A. Nigg
Ralf B. Schittenhelm and Sebastian Heeger have equal contributions.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License ( https://creativecommons.org/licenses/by-nc/2.0 ), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Schittenhelm, R.B., Heeger, S., Althoff, F. et al. Spatial organization of a ubiquitous eukaryotic kinetochore protein network in Drosophila chromosomes. Chromosoma 116, 385–402 (2007). https://doi.org/10.1007/s00412-007-0103-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-007-0103-y