Abstract
Despite the widespread use of the SH-SY5Y human neuroblastoma cell line in modeling human neurons in vitro, protocols for growth, differentiation and experimentation differ considerably across the literature. Many studies fully differentiate SH-SY5Y cells before experimentation, to investigate plasticity measures in a mature, human neuronal-like cell model. Prior to experimentation, serum is often removed from cell culture media, to arrest the cell growth cycle and synchronize cells. However, the exact effect of this serum removal before experimentation on mature, differentiated SH-SY5Y cells has not yet been described. In studies using differentiated SH-SY5Y cells, any effect of serum removal on plasticity markers may influence results. The aim of the current study was to systematically characterize, in differentiated, neuronal-like SH-SY5Y cells, the potentially confounding effects of complete serum removal in terms of morphological and gene expression markers of plasticity. We measured changes in commonly used morphological markers and in genes related to neuroplasticity and synaptogenesis, particularly in the BDNF-TrkB signaling pathway. We found that complete serum removal from already differentiated SH-SY5Y cells increases neurite length, neurite branching, and the proportion of cells with a primary neurite, as well as proportion of βIII-Tubulin and MAP2 expressing cells. Gene expression results also indicate increased expression of PSD95 and NTRK2 expression 24 h after serum removal. We conclude that serum deprivation in differentiated SH-SY5Y cells affects morphology and gene expression and can potentially confound plasticity-related outcome measures, having significant implications for experimental design in studies using differentiated SH-SY5Y cells as a model of human neurons.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
SH-SY5Y cells are a human neuroblastoma-derived cell line used to model human neurons in vitro. The original cells were derived from a bone marrow biopsy in 1970, and were cloned to produce the neuron-like SH-SY5Y cells that are used in a wide range of research applications today (Biedler et al. 1978). These cells synthesize various neurotransmitters, express neural markers, and can be further differentiated in vitro to a mature human neuronal phenotype (Jahn et al. 2017; Encinas et al. 2000, 1999; Shipley et al. 2016). Once differentiated, SH-SY5Y cells express a catecholaminergic phenotype, with the potential to synthesize both dopamine and noradrenaline (Krishna et al. 2014). They can be used to study synapse modifications and functional cellular activity with live calcium imaging or electrophysiology (Santillo et al. 2014; Toselli et al. 1996; Jahn et al. 2017). They are often used as a cell model for Parkinson’s Disease (Xicoy et al. 2017), as well as Alzheimer’s Disease (Agholme et al. 2010), neuropathogenesis of viruses (Christensen et al. 2011), screening for neurotropic properties of pharmaceuticals (Henkel et al. 2008; Xu et al. 2019), neurotoxicity (De Simone et al. 2018; Forster et al. 2016), and even as a multicellular 3D culture (Cui et al. 2017; Kapalczynska et al. 2018).
With the widespread use of this cell line to study human neuron synapse activity and neuronal plasticity in vitro, it is important to understand the effects of cell handling, such as the removal of serum before experimental manipulation in fully differentiated cells.
SH-SY5Y cells are grown in a basic medium containing Dulbecco’s Modified Eagle’s- Medium (DMEM), glucose, antibiotics, and supplemented with 10–20% Fetal Bovine Serum (FBS)(Xicoy et al. 2017). The use of FBS in culture media to promote growth of cells and to maintain tissues in vitro was introduced in 1958 (Puck et al. 1958). This serum supplementation is vital for the growth and maintenance of cell lines, as it contains many crucial proteins, vitamins, hormones and growth factors important for cell survival and proliferation (van der Valk et al. 2018).
To induce differentiation of SH-SY5Y cells to a more mature neuronal phenotype, the serum concentration is commonly reduced to 1% or 3%, along with the addition of retinoic acid (Xicoy et al. 2017; Encinas et al. 2000). After 5–20 days, depending on the differentiation protocol, the cells reach their maximum differentiation state (Encinas et al. 2000; Shipley et al. 2016; Jahn et al. 2017). Prior to experimental manipulation, e.g. exposure to potential pharmaceutical compounds, serum is often completely removed from the cultures. This is done to ensure all cells are in the same growth cycle phase before manipulation (Langan and Chou 2011), and to prevent confounding effects of the myriad of proteins and other molecular factors present in serum, which differ by serum batch and therefore introduce phenotypic variations in cell cultures (van der Valk et al. 2010). Serum components may also mask certain intrinsic growth factor (e.g. brain-derived neurotrophic factor, BDNF) effects, therefore, serum may be removed to assess the effects of BDNF in the absence of external growth factors (Zainullina et al. 2019).
Despite the common practice of serum removal before experimental manipulation in already differentiated SH-SY5Y cells, the effects of removing serum from culture media on plasticity-related gene expression and morphology markers have not yet been examined. Understanding the effects of serum removal is essential in standardizing pre-experimental protocols. If serum removal has strong effects on gene and morphological markers in already differentiated cells, any effect of experimentation may be confounded.
Here, we aim to systematically characterize the effects of completely removing serum from differentiated SH-SY5Y culture media on gene expression markers of plasticity, specifically related to an important pathway in synaptic plasticity and long-term potentiation, the BDNF-TrkB signaling pathway (Kowianski et al. 2018; Minichiello 2009; Yoshii and Constantine-Paton 2010; Andero et al. 2014; Leal et al. 2017; Niculescu et al. 2018). We also investigated the effects of serum removal on cytoskeletal markers of neuron morphology by visualizing changes in MAP2 and βIII-Tubulin.
Methods
Cell Culture
Cells were obtained from ATCC® (CRL2266™, RRID:CVCL_0019) and were maintained and expanded according to the provided protocol. For experiments, cells were not used above passage 26.
Undifferentiated cells were cultured in DMEM/ F12, GlutaMAX™ Supplement (GibcoTM, Thermo Scientific) supplemented with 10% heat inactivated Fetal Bovine Serum (FBS, Merck), 1% penicillin–streptomycin (P/S) and 1% L-Glutamate at 37 °C and 5% CO2, and split at 80–90% confluency.
Differentiation
All cells were fully differentiated to a mature neuronal-like state before experimentation. For differentiation, cells were plated in 6-well culture plates (Greiner CELLSTAR®, Merck) at approximately 2.4 × 104 cells per well. Serum concentration was decreased to 3% FBS three days prior to the addition of 10 µM retinoic acid (RA; Sigma-Aldrich, R2625). A stock solution of RA was prepared in dimethylsulphoxide (DMSO; Sigma-Aldrich, 41,640) at 10 mM, and stored at −20 °C until dilution in cell culture media to a final concentration of 10 µM. Starting from the day RA was added, medium with 3% FBS supplementation was replaced every two days for a total of ten days.
Serum Deprivation
Differentiated SH-SY5Y cells were used for serum removal experiments. Medium containing 3% serum (FBS) was removed, and the cell surface was rinsed with PBS (warmed to 37 °C) to remove all remaining serum. Next, medium without supplemented serum (0% FBS), or medium with serum (3% FBS) was added for 1 h, 3 h, 6 h or 24 h. In total there were 8 different conditions; serum and no serum for each of the 4 time points (1 h, 3 h, 6 h, 24 h).
Microscopy
Cells were grown on 12 mm glass coverslips (VWR, 631-1577) coated with 100 µg/mL Poly-L-Ornithine (Sigma, P4957) and 1 µg/mL Laminin (Sigma, L2020) and differentiated as described above.
At the specified collection time points following complete serum removal, cells were washed in PBS (warmed to 37 °C), and fixed for 10 min in cold 4% paraformaldehyde. To stop fixation, cells were washed 3 times 5 min in cold PBS and stored in PBS at 4 °C for a maximum of two days before antibody incubation. Cells were then blocked in PBS-Tween 20, prepared with 0.2% Tween-20, and 10% donkey serum. Antibodies for visualizing neurite outgrowth (βIII-Tubulin; Cell Signaling, Cat #5568S, RRID:AB_10694505), and dendrites (MAP2; Sigma, Cat #M2320, AB_609904) were used. Both markers were chosen as they are often combined to capture all neuronal processes (Paik et al. 2019), and they have consistently been used as markers of differentiation in experiments with SH-SY5Y cells (Jahn et al. 2017; Shipley et al. 2016; Encinas et al. 2000; Kovalevich and Langford 2013; Paik et al. 2019). Following primary antibody incubation, cells were washed in alternating PBS and PBS-Tween 20 and incubated with secondary antibodies donkey anti-rabbit Alexa 488 (Invitrogen, Cat #A-21206 RRID:AB_141708), donkey anti-mouse Alexa 594 (Invitrogen, Cat #A-21203, RRID:AB_141633), and cell nuclei were stained with DAPI (CarlRoth, Cat #6843.3). Following secondary antibody incubation, coverslips were washed in cold PBS and mounted on glass microscope slides. Fluorescence imaging was done with the Olympus BX51WI microscope and disc spinning unit. Pictures were taken using the 20X objective lens. Micro-Manager software (Edelstein et al. 2014)(RRID:SCR_016865) was used to collect images. Further details on the primary and secondary antibody dilutions as well as microscope exposure times can be found in the Supplementary Material (Table S1 and S2).
qRT-PCR
Cells in 6-well plates were first rinsed with PBS at 37 °C and then kept on ice for the rest of the extraction. RNA was extracted with TRIzol (Invitrogen, 15,596,026) according to the manufacturer’s protocol. RNA concentration was determined using the NanoDrop™ spectrophotometer, and cDNA was synthesized using RevertAid H Minus First Strand cDNA Synthesis Kit (Thermo Scientific, K1632). RNA was stored at −80 °C and cDNA at −20 °C. Three cell culture replicates were collected per time point, per condition.
Primers for qPCR were designed using NCBI gene reference database and Primer-BLAST (National Library of Medicine). The following primers were analyzed: Activity Regulated Cytoskeleton associated protein (ARC,Gene ID: 23,237), Early Response 1 (EGR1, Gene ID: 1958), cAMP Responsive Element Binding Protein 1 (CREB1, Gene ID: 1385), B-cell lymphoma 2 (BCL2, Gene ID: 596), BCL2- Associated X (BAX, Gene ID: 581), Brain Derived Neurotrophic Factor (BDNF, Gene ID: 627), Neurotrophic Receptor Tyrosine Kinase 2 (NTRK2, GeneID: 4915), Discs Large MAGUK scaffold protein 4 (DLG4 also known as PSD95, GeneID:1742), Synaptophysin (SYP, GeneID:6855), in conjunction with three House Keeping Genes (HKG’s): Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH, GeneID:2597), TATA-box Binding Protein (TBP,GeneID:6908), Peptidylprolyl Isomerase B (PPiB, GeneID:5479). Primer sequences can be found in Supplementary Material Table S3. Primers at 600 nM concentration were mixed with Fast Start Universal Sybr Green Master ROX (Roche,491,385,001). Samples were run in 384-well qPCR plates (Roche,4TI-0382), using the LightCycler 480 Real-Time PCR system (Roche Life Sciences). qPCR program details are described in Supplementary Material Table S4.
Analysis
Microscopy
Image processing and analysis was done in Fiji (ImageJ version 1.52i, RRID:SCR_002285) (Schindelin et al. 2012). A DAPI nucleus staining was used to count total cells in each image. Neurite length and branching was measured in the 488 (βIII-Tubulin) channel, using the segmented line tool at 20X magnification. Per condition 70–100 neurites were measured. Typically, in differentiated SH-SY5Y cultures, many cells have only one or no neurites. Therefore, from each cell, we measured the length of the primary neurite, defined as the single neurite, or the longest neurite for cells having more than one neurite. An example of tracings of primary neurites as well as cells without neurite extensions can be seen in Fig. 2a. The NeuronJ plugin (Meijering et al. 2004) was used to quantify neurite length and neurite branching. For each image, primary neurites and branches were semi-automatically traced, and manually labeled as either primary neurites, or branches. The number of branches were divided by the total neurons (counted with DAPI), to give the number of branches per neuron in each image. An example neuron with branching can be seen in Fig. 2b. To identify whether the proportion of total cells with a primary extension changes due to serum deprivation, the number of primary neurites was also divided by total number of neurons (as counted with DAPI).
Total fluorescence of βIII-Tubulin staining was quantified using the 488 channel. First, the fluorescence threshold was set with the minimum intensity as the maximum background intensity. The total fluorescence intensity in the image was then measured, and corrected for cell area by dividing by the total area of cells in the 488 channel.
βIII-Tubulin stained cells were counted manually by setting the brightness contrast settings to 834 (min) and 7474 (max). Cells with visible green neurites were counted. MAP2 was manually counted in the 594 channel, at brightness contrast settings of 596 (min) and 6007 (max). These cell counts were divided by the total cells to calculate the proportion of βIII Tubulin or MAP2 cells.
Gene Expression
A standard curve was used to calculate relative concentrations of gene expression per gene. An average of technical duplicates was made, and normalized to the average of 3 HKGs (TBP, PPiB, GAPDH). Analyses were performed with LightCycler 480 software version 1.5.1.62 (Roche Life Sciences) and Excel.
Statistics
Statistical analysis and graphs were made with Prism 5 (Graphpad Software, USA, RRID:SCR_002798) and IBM Statistics 24 (SPSS for windows version 24.0, Armonk, NY:IBM Corp). Data collected from 2 independent experiments were pooled for statistical analysis. This resulted in a total of 4–14 images per condition being included in statistical analysis. For analysis of neurite length, an average neurite length per image was calculated from 70 to 100 neurites, for a total of 8 images per condition included in the 2-way ANOVA. For the analysis of primary neurites per neuron and neurite branching per neuron, the number of primary neurites in an image (between 30 and 100 per image) were divided by the total neurons in the image (between 210 and 250 cells per image), with 4–6 images per condition included in the 2-way ANOVA. For analysis of βIII-Tubulin Immunoreactivity, a total of 10–14 images per condition were included in the 2-way ANOVA. A 2-way ANOVA with factors Serum (serum, no serum) and Time (1,3,6, 24 h) was used for all comparisons of microscopy quantification and HKG normalized expression values. Bonferroni-corrected post-hoc tests were done in the case of significant interaction events. Reported results are mean ± standard error of the mean. Figures show bar graphs of the HKG normalized mean expression values; error bars are standard errors of the mean.
Results
Differentiation
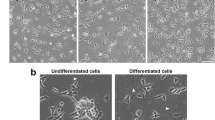
Differentiation was verified as explained previously (Thomson et al. 2020). Representative images comparing undifferentiated and differentiated cells can be seen in Fig. 1.
Microscopy
Full statistical results of main effects (Time, Serum) and Time x Serum interaction effects for each parameter (Neurite Length, Neurite Branching, Primary Neurites, βIII-Tubulin immunoreactivity, and βIII-Tubulin and MAP2 positive cells) can be found in Supplementary Material Table S5. In case of significant main or interaction effects, the p-value of Bonferroni-corrected post-hoc tests are reported below. An example of the parameters measured can be seen in Fig. 2. An example of the morphology of cells immediately following serum removal can be seen in Supplementary Fig. 2.
Neurite Length
We found a significant effect of Serum (p < 0.0001), but no significant effect of Time (p = 0.382), or Time x Serum interaction (p = 0.338) on neurite length. Serum deprived neurons had significantly longer outgrowths than neurons with serum at 3 h (54.93 ± 20.22 μm vs. 45.43 ± 17.76 μm, p = 0.03) and at 24 h (65.53 ± 27.74 μm vs. 46.89 ± 18.24 μm, p < 0.0001) (Fig. 3a).
Neurite Branching
There was a significant effect of Time (p = 0.0105) and Serum (p < 0.0001), but not a significant Time × Serum interaction (p = 0.062) on neurite branching. Serum deprived neurons had significantly more branches per neuron at 3 h (0.058 ± 0.008 vs. 0.011 ± 0.003, p < 0.0001), 6 h (0.041 ± 0.007 vs. 0.015 ± 0.003, p < 0.028), and at 24 h (0.033 ± 0.008 vs. 0.0045 ± 0.003, p < 0.028) (Fig. 3b).
Primary Neurites
Similarly, we found a significant effect of Time (p = 0.0368), Serum (p = 0.0006), but not a significant Time x Serum interaction (p = 0.109). At 3 h, serum deprived neurons showed a greater proportion with a primary neurite (0.45 ± 0.034 vs 0.25 ± 0.020, p = 0.001) (Fig. 3c).
βIII-Tubulin Immunoreactivity
A significant effect of Serum (p = 0.0006) and a Time x Serum interaction (p = 0.0035) was found, but no effect of Time (p = 0.14) on βIII-Tubulin immunoreactivity. There was a significant increase in βIII-Tubulin immunoreactivity in the serum deprived cells at 3 h (120.23 ± 25.58 vs. 37.03 ± 4.96, p < 0.01) and 6 h (110.49 ± 28.55 vs. 22.94 ± 3.03, p < 0.01) (Fig. 3d).
βIII-Tubulin and MAP2 Positive Cells
There was a significant effect of Time (p < 0.0001), Serum (p < 0.0001), and a Time × Serum interaction (p = 0.0012), on cells expressing βIII-Tubulin after serum deprivation. Serum deprived neurons showed a significant increase in the percentage of βIII-Tubulin expressing cells at 3 h (42.04 ± 4.92% vs. 23.16 ± 3.78%, p = 0.009) and 6 h (30.80 ± 2.77% vs. 17.40 ± 2.21%, p = 0.023), and 24 h (26.69 ± 2.41 vs. 8.90 ± 2.35, p = 0.00012) (Fig. 3e).
There was also a significant effect of Time (p < 0.0001), Serum (p < 0.0001), and a Time x Serum interaction (p = 0.0011), on percentage of cells expressing MAP2 after serum deprivation. Serum deprived cells also showed a significant increase in percentage MAP2 expressing cells at 3 h (29.08 ± 5.02% vs. 15.30 ± 2.28%, p < 0.01), 6 h (23.65 ± 2.12% vs. 12.51 ± 1.87%, p < 0.05), and 24 h (19.85 ± 2.50 vs. 7.27 ± 2.21, p < 0.01) (Fig. 3f).
Gene Expression
We were most interested in gene expression changes following serum deprivation, specifically in genes related to IEG expression (ARC, EGR1), apoptosis (BCL2, BAX), plasticity (BDNF, NTRK2, CREB1) and synaptogenesis (PSD95, SYP). Full statistical results of main (Time, Serum) and Time x Serum interaction effects can be found in Supplementary Material Table S6. In case of significant main or interaction effects, p values and Bonferroni-corrected post hoc tests are reported in the text. Graphs show mean HKG normalized expression levels for each condition, error bars are standard error of the mean.
Immediate Early Gene Expression
We measured the expression of IEG’s ARC and EGR1, finding high expression levels in both genes in the 1 h serum condition only. Expression levels were low in all other samples (3 h, 6 h and 24 h), many of which were too low to detect (Ct value ≥ 34). Gene expression results can be found in Supplementary Material and Supplementary Fig. 1.
Apoptosis Markers
BCL2 Expression
We also found no significant main effects of Time (p = 0. 362), Serum (p = 0.618), or Time × Serum interaction (p = 0.216) on BCL2 expression (Fig. 4a).
BAX Expression
We found no significant main effect of Time (p = 0.169), Serum (p = 0.380), or Time × Serum interaction (p = 0.228) on BAX expression (Fig. 4b).
Expression of BDNF Signaling
BDNF Expression
We found a significant effect of Time (p < 0.001), Serum (p < 0.001) and Time × Serum interaction (p < 0.001) on BDNF expression. There was a significant decrease in BDNF expression in the serum deprived cells at 1 h (p < 0.0001), 3 h (p < 0.0001), and 6 h (p < 0.0001). Compared to time-matched serum controls, serum deprived cells express 25.71 ± 5.76% BDNF at 1 h, 8.07 ± 1.35% at 3 h, 12.01 ± 0.61% at 6 h and 94.58 ± 5.89% at 24 h (Fig. 5a).
NTRK2 Expression
There was also a significant effect of Time (p = 0. 008), Serum (p = 0.006), and Time × Serum interaction (p = 0.021) on NTRK2 expression. There was a significant increase in expression of NTRK2 mRNA at 24 h (p < 0.01), with serum deprived cells expressing 242.11 ± 33.00% of the NTRK2 expressed in serum controls (Fig. 5b).
CREB Expression
There was no significant main effect of Time (p = 0. 393) or Serum (p = 0.942), on CREB expression. However, there was a slight Time × Serum interaction effect (p = 0.043). Initially, there is a decrease in CREB expression in serum deprived cells at 1 h (to 65.25 ± 1.26% serum controls), at 6 h this is reversed (164.71 ± 30.31% serum controls) (Fig. 5c). None of these time points are significant in Bonferroni-corrected post hoc tests.
Synaptogenesis Genes
PSD95 Expression
There was a significant effect of Time (p = 0.001), and a Time × Serum interaction (p = 0.020), but no effect of Serum (p = 0.153), on PSD95 expression. There was a significant increase in expression in the serum-deprived cells at 24 h (p < 0.01).Serum-deprived cells express 192.85 ± 17.78% of the serum controls at 24 h (Fig. 6a).
SYP Expression
Again, we found a significant effect of Time (p = 0.017), but no effect of Serum (p = 0.575). We found a trend towards a significant Time × Serum interaction effect (p = 0.059). (Fig. 6b).
Discussion
In this study, we aimed to systematically characterize, in fully differentiated SH-SY5Y cells, the effects of complete serum removal on several morphological and gene expression markers of plasticity. We found that serum removal, over 24 h, increased primary neurite length as well as neurite branching when compared to serum controls. Serum deprived neurons also showed higher levels of βIII-Tubulin immunoreactivity, and a greater proportion of βIII-Tubulin- and MAP2-positive neurons. MAP2 is mainly localized in mature dendrites (Dehmelt and Halpain 2005), and βIII-Tubulin is a widely used neuronal maturity marker (Katsetos et al. 1993). These findings suggest that in fully differentiated cells, complete serum removal may promote additional plasticity-like effects. This can be seen as early as 3 h following removal of serum, and lasts at least 24 h.
We also found that complete serum removal has a specific effect on the expression of several genes involved in BDNF-TrkB signaling and synaptogenesis. Serum deprivation resulted in a significant increase in the expression of NTRK2, PSD95 and SYP over time, with the strongest effect on the expression of NTRK2 and PSD95 mRNA at 24 h following deprivation. NTRK2, the gene coding for the TrkB receptor, has been shown to be important in activity dependent plasticity leading to long term potentiation (LTP) (Minichiello 2009; Minichiello et al. 1999). PSD-95 is an important scaffolding protein, regulating the strength of excitatory synapses (Chen et al. 2015), and the SYP gene codes for synaptophysin, an important protein involved in neurotransmitter release (Südhof et al. 1987; Arthur and Stowell 2007). An increase in the expression of NTRK2, PSD95 and SYP mRNA over time in serum-deprived cells therefore aligns with our morphology results. Our results suggest that complete serum removal induces an increased expression of genes and morphological markers of plasticity and synaptic strength, potentially confounding experiments interested in these outcome measures.
While these observations are in line with the differentiation-inducing effect of serum deprivation (Encinas et al. 2000; Jahn et al. 2017; Shipley et al. 2016; Kaplan et al. 1993), these cells are already fully differentiated, therefore the additional changes in morphological markers that we present here may indicate additional, confounding plasticity effects. Indeed, systematic transcriptomic profiling SH-SY5Y cells has identified NTRK2 as well as many genes involved in neurogenesis and cytoskeletal reorganization as upregulated in differentiated compared to undifferentiated cells (Pezzini et al. 2017). However, once cells have been differentiated, the expression of these genes is stable over time; in contrast to the serum removal effects we report here. This semi-acute increase in plasticity-related gene expression and morphological markers is problematic in studies using these genes or morphological plasticity markers as outcome measures.
Interestingly, we also report a strong effect of serum on the expression of BDNF, ARC and EGR1. BDNF expression increased in the in the serum control cells after 1, 3 and 6 h, returning to low expression levels at 24 h. The serum control cells underwent a regular medium change, including a PBS wash step. This increase in BDNF mRNA in the serum condition is surprising, but may be related to the addition of fresh medium and serum. In serum-deprived cells, this temporary increase is absent, likely due to the disruption of growth and protein production as a consequence of serum withdrawal (Inoue et al. 1996; Irie et al. 1999; Ozturk et al. 2003; Ozturk and Palsson 1991). We also report an increase in expression of the immediate early genes ARC and EGR1 in the serum condition at 1 h after PBS wash and serum replacement. The removal and re-addition of serum could have induced an immediate but transient increase in the expression of ARC and EGR1 mRNA, in line with the expected expression pattern of an immediate early gene (Lyford et al. 1995; Schratt et al. 2001).
We did not find any effects of serum deprivation on the expression of genes linked to apoptosis, BAX and BCL2. Encinas et al. (2000) showed that SH-SY5Y cells show signs of apoptosis 6 and 24 h after serum removal as measured by caspase activity and TUNEL assay (Encinas et al. 2000). Encinas et al. (2000) used cells that were treated with RA for only five days and in medium containing 15% FBS. The shock of serum removal in the not-fully differentiated cells is likely much stronger compared to our protocol, and may explain the different finding. Based on the gene expression markers in our experiments, we cannot confirm that serum starvation influences apoptotic processes after 24 h in in fully differentiated SH-SY5Y cells.
Conclusion
Despite being common practice to remove serum from the culture medium of already differentiated SH-SY5Y cultures before experimentation, the effects on morphology and gene expression had not been systematically characterized. Here, we show that complete serum deprivation has an effect on commonly used morphological and gene expression markers of cellular and synaptic plasticity in differentiated SH-SY5Y cells, and may thus confound results when examining plasticity-related outcome measures. For future research involving differentiated SH-SY5Y cells as a model of human neural plasticity, our findings provide some key considerations for experimental design. Studies interested in measuring plasticity effects in differentiated SH-SY5Y cells should either refrain from complete serum deprivation 24 h before experimentation, or include appropriate controls, e.g. cells which were not serum deprived, to confirm serum deprivation had no confounding effects on outcome measures.
Data Availability
All data available upon reasonable request.
Abbreviations
- FBS:
-
Fetal Bovine Serum
- RA:
-
Retinoic Acid
- MAP2:
-
Microtubule Associated Protein 2
- ARC:
-
Activity Regulated Cytoskeleton associated protein
- EGR1:
-
Early Growth Response 1
- BCL2:
-
B-cell Lymphoma 2
- BAX:
-
BCL2- Associated X
- BDNF:
-
Brain-Derived Neurotrophic Factor
- NTRK2:
-
Neurotrophic Receptor Tyrosine Kinase 2
- CREB1:
-
CAMP Responsive Element Binding Protein 1
- PSD95:
-
Discs Large MAGUK scaffold protein 4, DLG4
- SYP:
-
Synaptophysin
- TBP:
-
TATA-Box Binding Protein
- GAPDH:
-
Glyceraldehyde 3-Phosphate Dehydrogenase
- PPiB:
-
Peptidylprolyl Isomerase B
References
Agholme L, Lindstrom T, Kagedal K, Marcusson J, Hallbeck M (2010) An in vitro model for neuroscience: differentiation of SH-SY5Y cells into cells with morphological and biochemical characteristics of mature neurons. J Alzheimers Dis 20(4):1069–1082. https://doi.org/10.3233/JAD-2010-091363
Andero R, Choi DC, Ressler KJ (2014) BDNF-TrkB receptor regulation of distributed adult neural plasticity, memory formation and psychiatric disorders. Prog Mol Biol Transl 122:169–192. https://doi.org/10.1016/B978-0-12-420170-5.00006-4
Arthur CP, Stowell MHB (2007) Structure of synaptophysin: a hexameric MARVEL-domain channel protein. Structure 15(6):707–714. https://doi.org/10.1016/j.str.2007.04.011
Biedler JL, Roffler-Tarlov S, Schachner M, Freedman LS (1978) Multiple neurotransmitter synthesis by human neuroblastoma cell lines and clones. Can Res 38(11 Pt 1):3751–3757
Chen X, Levy JM, Hou A, Winters C, Azzam R, Sousa AA, Leapman RD, Nicoll RA, Reese TS (2015) PSD-95 family MAGUKs are essential for anchoring AMPA and NMDA receptor complexes at the postsynaptic density. Proc Natl Acad Sci U S A 112(50):E6983-6992. https://doi.org/10.1073/pnas.1517045112
Christensen J, Steain M, Slobedman B, Abendroth A (2011) Differentiated neuroblastoma cells provide a highly efficient model for studies of productive varicella-zoster virus infection of neuronal cells. J Virol 85(16):8436–8442. https://doi.org/10.1128/JVI.00515-11
Cui X, Hartanto Y, Zhang H (2017) Advances in multicellular spheroids formation. J R Soc Interface 14(127):20160877. https://doi.org/10.1098/rsif.2016.0877
De Simone U, Roccio M, Gribaldo L, Spinillo A, Caloni F, Coccini T (2018) Human 3D cultures as models for evaluating magnetic nanoparticle CNS cytotoxicity after short- and repeated long-term exposure. Int J Mol Sci. https://doi.org/10.3390/ijms19071993
Dehmelt L, Halpain S (2005) The MAP2/Tau family of microtubule-associated proteins. Genome Biol 6(1):204. https://doi.org/10.1186/gb-2004-6-1-204
Edelstein AD, Tsuchida MA, Amodaj N, Pinkard H, Vale RD, Stuurman N (2014) Advanced methods of microscope control using μManager software. https://doi.org/10.14440/jbm.2014.36
Encinas M, Iglesias M, Llecha N, Comella JX (1999) Extracellular-regulated kinases and phosphatidylinositol 3-kinase are involved in brain-derived neurotrophic factor-mediated survival and neuritogenesis of the neuroblastoma cell line SH-SY5Y. J Neurochem 73:1409–1421. https://doi.org/10.1046/j.1471-4159.1999.0731409.x
Encinas M, Iglesias M, Liu Y, Wang H, Muhaisen A, Cena V, Gallego C, Comella JX (2000) Sequential treatment of SH-SY5Y cells with retinoic acid and brain-derived neurotrophic factor gives rise to fully differentiated, neurotrophic factor-dependent, human neuron-like cells. J Neurochem 75(3):991–1003. https://doi.org/10.1046/j.1471-4159.2000.0750991.x
Forster JI, Koglsberger S, Trefois C, Boyd O, Baumuratov AS, Buck L, Balling R, Antony PM (2016) Characterization of Differentiated SH-SY5Y as Neuronal Screening Model Reveals Increased Oxidative Vulnerability. J Biomol Screen 21(5):496–509. https://doi.org/10.1177/1087057115625190
Henkel AW, Sperling W, Rotter A, Reulbach U, Reichardt C, Bonsch D, Maler JM, Kornhuber J, Wiltfang J (2008) Antidepressant drugs modulate growth factors in cultured cells. BMC Pharmacol 8:6. https://doi.org/10.1186/1471-2210-8-6
Inoue Y, Lopez LB, Kawamoto S, Arita N, Teruya K, Seki K, Shoji M, Kamei M, Hashizume S, Shiozawa Y, Tachibana H, Ohashi H, Yasumoto K, Nagashima A, Nakahashi H, Suzuki T, Imai T, Nomoto K, Takenoyama M, Katakura Y, Shirahata S (1996) Production of recombinant human monoclonal antibody using ras-amplified BHK-21 cells in a protein-free medium. Biosci Biotechnol Biochem 60(5):811–817. https://doi.org/10.1271/bbb.60.811
Irie A, Lee KE, Kadowaki K, Toda K, Yamada Y (1999) Elevation of serum and urine tumor necrosis factor levels after transurethral resection of the prostate. Nihon Hinyokika Gakkai Zasshi 90(4):502–508. https://doi.org/10.5980/jpnjurol1989.90.502
Jahn K, Wieltsch C, Blumer N, Mehlich M, Pathak H, Khan AQ, Hildebrandt H, Frieling H (2017) A cell culture model for investigation of synapse influenceability: epigenetics, expression and function of gene targets important for synapse formation and preservation in SH-SY5Y neuroblastoma cells differentiated by retinoic acid. J Neural Transm (Vienna) 124(11):1341–1367. https://doi.org/10.1007/s00702-017-1769-9
Kapalczynska M, Kolenda T, Przybyla W, Zajaczkowska M, Teresiak A, Filas V, Ibbs M, Blizniak R, Luczewski L, Lamperska K (2018) 2D and 3D cell cultures - a comparison of different types of cancer cell cultures. Arch Med Sci 14(4):910–919. https://doi.org/10.5114/aoms.2016.63743
Kaplan DR, Matsumoto K, Lucarelli E, Thiele CJ (1993) Induction of TrkB by retinoic acid mediates biologic responsiveness to BDNF and differentiation of human neuroblastoma cells. Eukaryotic Signal Transduction Group Neuron 11(2):321–331. https://doi.org/10.1016/0896-6273(93)90187-v
Katsetos CD, Frankfurter A, Christakos S, Mancall EL, Vlachos IN, Urich H (1993) Differential localization of class III, beta-tubulin isotype and calbindin-D28k defines distinct neuronal types in the developing human cerebellar cortex. J Neuropathol Exp Neurol 52:655–666. https://doi.org/10.1097/00005072-199311000-00013
Kovalevich J, Langford D (2013) Considerations for the use of SH-SY5Y neuroblastoma cells in neurobiology. Methods Mol Biol 1078:9–21. https://doi.org/10.1007/978-1-62703-640-5_2
Kowiański P, Lietzau G, Czuba E, Waśkow M, Steliga A, Moryś J (2018) BDNF: a key factor with multipotent impact on brain signaling and synaptic plasticity. Cell Mol Neurobiol 38(3):579–593
Krishna A, Biryukov M, Trefois C, Antony PM, Hussong R, Lin J, Heinaniemi M, Glusman G, Koglsberger S, Boyd O, van den Berg BH, Linke D, Huang D, Wang K, Hood L, Tholey A, Schneider R, Galas DJ, Balling R, May P (2014) Systems genomics evaluation of the SH-SY5Y neuroblastoma cell line as a model for Parkinson’s disease. BMC Genomics 15:1154. https://doi.org/10.1186/1471-2164-15-1154
Langan TJ, Chou RC (2011) Synchronization of mammalian cell cultures by serum deprivation. Methods Mol Biol 761:75–83. https://doi.org/10.1007/978-1-61779-182-6_5
Leal G, Bramham CR, Duarte CB (2017) BDNF and hippocampal synaptic plasticity. Vitam Horm 104:153–195. https://doi.org/10.1016/bs.vh.2016.10.004
Lyford GL, Yamagata K, Kaufmann WE, Barnes CA, Sanders LK, Copeland NG, Gilbert DJ, Jenkins NA, Lanahan AA, Worley PF (1995) Arc, a growth factor and activity-regulated gene, encodes a novel cytoskeleton-associated protein that is enriched in neuronal dendrites. Neuron 14(2):433–445. https://doi.org/10.1016/0896-6273(95)90299-6
Meijering E, Jacob M, Sarria JC, Steiner P, Hirling H, Unser M (2004) Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A 58(2):167–176. https://doi.org/10.1002/cyto.a.20022
Minichiello L (2009) TrkB signalling pathways in LTP and learning. Nat Rev Neurosci 10(12):850–860. https://doi.org/10.1038/nrn2738
Minichiello L, Korte M, Wolfer D, Kuhn R, Unsicker K, Cestari V, Rossi-Arnaud C, Lipp HP, Bonhoeffer T, Klein R (1999) Essential role for TrkB receptors in hippocampus-mediated learning. Neuron 24(2):401–414. https://doi.org/10.1016/s0896-6273(00)80853-3
Niculescu D, Michaelsen-Preusse K, Guner U, van Dorland R, Wierenga CJ, Lohmann C (2018) A BDNF-mediated push-pull plasticity mechanism for synaptic clustering. Cell Rep 24(8):2063–2074. https://doi.org/10.1016/j.celrep.2018.07.073
Ozturk S, Kaseko G, Mahaworasilpa T, Coster HG (2003) Adaptation of cell lines to serum-free culture medium. Hybrid Hybridomics 22(4):267–272. https://doi.org/10.1089/153685903322329009
Ozturk SS, Palsson BO (1991) Physiological changes during the adaptation of hybridoma cells to low serum and serum-free media. Biotechnol Bioeng 37(1):35–46. https://doi.org/10.1002/bit.260370107
Paik S, Somvanshi RK, Kumar U (2019) Somatostatin-mediated changes in microtubule-associated proteins and retinoic acid-induced neurite outgrowth in SH-SY5Y cells. J Mol Neurosci 68(1):120–134. https://doi.org/10.1007/s12031-019-01291-2
Pezzini F, Bettinetti L, Di Leva F, Bianchi M, Zoratti E, Carrozzo R, Santorelli FM, Delledonne M, Lalowski M, Simonati A (2017) Transcriptomic profiling discloses molecular and cellular events related to neuronal differentiation in SH-SY5Y neuroblastoma cells. Cell Mol Neurobiol 37(4):665–682. https://doi.org/10.1007/s10571-016-0403-y
Puck TT, Cieciura SJ, Robinson A (1958) Genetics of somatic mammalian cells. III. Long-term cultivation of euploid cells from human and animal subjects. J Exp Med 108(6):945–956. https://doi.org/10.1084/jem.108.6.945
Santillo S, Schiano Moriello A, Di Maio V (2014) Electrophysiological variability in the SH-SY5Y cellular line. Gen Physiol Biophys 33(1):121–129. https://doi.org/10.4149/gpb_2013071
Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9(7):676–682. https://doi.org/10.1038/nmeth.2019
Schratt G, Weinhold B, Lundberg AS, Schuck S, Berger J, Schwarz H, Weinberg RA, Rüther U, Nordheim A (2001) Serum response factor is required for immediate-early gene activation yet is dispensable for proliferation of embryonic stem cells. Mol Cell Biol 21(8):2933–2943. https://doi.org/10.1128/MCB.21.8.2933-2943.2001
Shipley MM, Mangold CA, Szpara ML (2016) Differentiation of the SH-SY5Y human neuroblastoma cell line. J Visualized Exp JVE 108:53193. https://doi.org/10.3791/53193
Südhof TC, Lottspeich F, Greengard P, Mehl E, Jahn R (1987) The cDNA and derived amino acid sequences for rat and human synaptophysin. Nucleic Acids Res 15(22):9607–9607. https://doi.org/10.1093/nar/15.22.9607
Thomson AC, Kenis G, Tielens S, de Graaf TA, Schuhmann T, Rutten BPF, Sack AT (2020) Transcranial magnetic stimulation-induced plasticity mechanisms: TMS-related gene expression and morphology changes in a human neuron-like cell model. Front Mol Neurosci. https://doi.org/10.3389/fnmol.2020.528396
Toselli M, Tosetti P, Taglietti V (1996) Functional changes in sodium conductances in the human neuroblastoma cell line SH-SY5Y during in vitro differentiation. J Neurophysiol 76(6):3920–3927. https://doi.org/10.1152/jn.1996.76.6.3920
van der Valk J, Bieback K, Buta C, Cochrane B, Dirks WG, Fu J, Hickman JJ, Hohensee C, Kolar R, Liebsch M, Pistollato F, Schulz M, Thieme D, Weber T, Wiest J, Winkler S, Gstraunthaler G (2018) Fetal Bovine Serum (FBS): past - present - future. ALTEX 35:99–118. https://doi.org/10.14573/altex.1705101
van der Valk J, Brunner D, De Smet K, Fex Svenningsen Å, Honegger P, Knudsen LE, Lindl T, Noraberg J, Price A, Scarino ML, Gstraunthaler G (2010) Optimization of chemically defined cell culture media – Replacing fetal bovine serum in mammalian in vitro methods. Toxicol In Vitro 24(4):1053–1063. https://doi.org/10.1016/j.tiv.2010.03.016
Xicoy H, Wieringa B, Martens GJ (2017) The SH-SY5Y cell line in Parkinson’s disease research: a systematic review. Mol Neurodegene 12(1):10. https://doi.org/10.1186/s13024-017-0149-0
Xu L, Su J, Guo L, Wang S, Deng X, Ma S (2019) Modulation of LPA1 receptor-mediated neuronal apoptosis by Saikosaponin-d: a target involved in depression. Neuropharmacology 155:150–161. https://doi.org/10.1016/j.neuropharm.2019.05.027
Yoshii A, Constantine-Paton M (2010) Postsynaptic BDNF-TrkB Signaling in synapse maturation, plasticity, and disease. Dev Neurobiol 70(5):304–322. https://doi.org/10.1002/dneu.20765
Zainullina LF, Gudasheva TA, Vakhitova YV, Seredenin SB (2019) Low-molecular-weight compound GSB-106 Mimics the cellular effects of BDNF after serum deprivation. Dokl Biochem Biophys 489(1):396–398. https://doi.org/10.1134/S1607672919060139
Funding
This work was supported by the Netherlands Organization for Scientific Research (NWO, Veni to T.A.G 451-13-024; Vidi to B.P.F.R. 917-18-336; Vici to A.T.S 453-15-008), and an internal grant from the Centre for Integrative Neuroscience at Maastricht University.
Author information
Authors and Affiliations
Contributions
Conceptualization, Alix Thomson and Gunter Kenis; Formal analysis, Alix Thomson; Funding acquisition, Alexander Sack and Bart Rutten; Investigation, Alix Thomson; Supervision, Teresa Schuhmann, Tom de Graaf, Alexander Sack, Bart Rutten and Gunter Kenis; Writing – original draft, Alix Thomson; Writing – review & editing, Teresa Schuhmann, Tom de Graaf, Alexander Sack, Bart Rutten and Gunter Kenis.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Thomson, A.C., Schuhmann, T., de Graaf, T.A. et al. The Effects of Serum Removal on Gene Expression and Morphological Plasticity Markers in Differentiated SH-SY5Y Cells. Cell Mol Neurobiol 42, 1829–1839 (2022). https://doi.org/10.1007/s10571-021-01062-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10571-021-01062-x