Abstract
Purpose
Caveolin-1 (CAV1) has been implicated in breast cancer oncogenesis and metastasis and may be a potential prognosticator, especially for non-distant events. CAV1 functions as a master regulator of membrane transport and cell signaling. Several CAV1 SNPs have been linked to multiple cancers, but the prognostic impact of CAV1 SNPs in breast cancer remains unclear. Here, we investigated CAV1 polymorphisms in relation to clinical outcomes in breast cancer.
Methods
A cohort of 1017 breast cancer patients (inclusion 2002–2012, Sweden) were genotyped using Oncoarray by Ilumina. Patients were followed for up to 15 years. Five out of six CAV1 SNPs (rs10256914, rs959173, rs3807989, rs3815412, and rs8713) passed quality control and were used for haplotype construction. CAV1 genotypes and haplotypes in relation to clinical outcomes were assessed with Cox regression and adjusted for potential confounders (age, tumor characteristics, and adjuvant treatments).
Results
Only one SNP was associated with lymph node status, no other SNPs or haplotypes were associated with tumor characteristics. The CAV1 rs3815412 CC genotype (5.8% of patients) was associated with increased risk of contralateral breast cancer, adjusted hazard ratio (HRadj) 4.26 (95% CI 1.86–9.73). Moreover, the TTACA haplotype (13% of patients) conferred an increased risk for locoregional recurrence HRadj 2.24 (95% CI 1.24–4.04). No other genotypes or haplotypes were associated with clinical outcome.
Conclusion
CAV1 polymorphisms were associated with increased risk for locoregional recurrence and contralateral breast cancer. These findings may identify patients that could derive benefit from more tailored treatment to prevent non-distant events, if confirmed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
Breast cancer remains a clinical challenge. Despite progress in treatment and diagnostics, some patients still relapse [1]. New prognostic and predictive biomarkers are needed to better tailor treatment to the individual patient [1, 2]. While many predictive and prognostic biomarkers exist in breast cancer [3,4,5], most focus on predicting distant metastasis. No specific biomarker exists for non-distant events, i.e., metachronous contralateral breast cancer or locoregional recurrence [6, 7]. Patients with a metachronous contralateral breast cancer or locoregional recurrence have a higher risk of developing distant metastasis and have worse survival compared to those without [8,9,10]. By convention, a metachronous contralateral breast cancer is considered a new primary tumor [11]. However, studies have shown that a subset of metachronous contralateral breast cancers represent a metastatic spread of the primary tumor [11, 12]. We previously reported that tumor-specific Caveolin-1 (CAV1) was prognostic for both contralateral breast cancer (CAV1 in malignant cells) and locoregional recurrence (CAV1 in stromal cells) [13]. Furthermore, host factors modulated how CAV1 in malignant and stromal cells affected prognosis [13]. It would, therefore, be of interest to further elucidate the role of CAV1 in breast cancer by studying CAV1 genotypes.
The CAV1 gene is located on human chromosome 7(7q31.1) and contains three exons, with the last exon encoding the bulk of the functional domains [14]. CAV1 is primarily located in cholesterol-rich plasma membrane raft domains (caveolae) and serves as a master regulator of cell signaling and transport, including drug internalization [15, 16]. CAV1 is most abundantly expressed in endothelial cells, fibroblasts, and adipocytes [14, 17]. CAV1 and caveolae have been implicated in several vital processes for breast cancer tumorigenesis and invasion, including inflammation, epithelial-mesenchymal transition, hypoxia response, and tumor–stroma interaction [15, 16, 18]. CAV1 has also been linked to radioresistance in various cancers through regulation of tyrosine kinase receptor membrane trafficking and thereby activating DNA repair mechanisms [18]. Moreover, CAV1 plays a crucial role in adipose tissue regulation, which is central to development of metabolic syndrome and obesity [19]. The loss of CAV1 in adipose tissue leads to an inability to store fat properly, leading to lipodystrophy, insulin resistance, hypertriglyceridemia, and metabolic syndrome [19, 20]. CAV1 deficiency in adipose tissue also leads to the recruitment of M2 macrophages [21] that promote tumorigenesis [22]. The role of CAV1 in obesity may be more prominent in women than in men [23]. Therefore, it would be of value to further explore adipose tissue regulators, such as CAV1 in breast cancer, considering the complex relationship between obesity and breast cancer [24, 25]. Specific CAV1 genotypes are associated with both fat distribution and waist circumference [26]. A meta-analysis showed associations between CAV1 SNPs and increased risk of breast cancer in Asian and Middle Eastern populations [27], and a similar association between CAV1 SNPs and gastrointestinal and urinary cancer risk has been reported [28, 29]. However, to our knowledge, there are no studies on the relationship between CAV1 genotypes and prognosis in breast cancer. Here, we investigated whether CAV1 genotypes and haplotypes impact prognosis, especially risk for metachronous contralateral breast cancer and locoregional recurrence, in primary breast cancer.
Materials and methods
Cohort description
BCblood is a population-based breast cancer cohort, consisting of patients with primary breast cancer operated at Skåne University Hospital, Lund. The study was approved by the Lund University Ethics Committee (Dnr 75-02, Dnr 37-08, Dnr 658-09, and amendments). All participants provided written informed consent. Inclusion of patients occurred between diagnosis and surgery. Only patients diagnosed with a first primary breast cancer and had not been diagnosed with cancer 10 years prior were included. At inclusion, a questionnaire regarding lifestyle and reproductive factors was answered, research nurses took anthropometric measurements and collected EDTA plasma for genotyping. Clinical data were obtained from medical records, pathology reports, and registries. After excluding patients with carcinoma in situ, preoperative treatment, and distant metastasis within 0.3 years of inclusion, and no available genotype, 1017 patients remained (inclusion October 2002 to June 2012, (Fig. 1). Last follow-up was June 30, 2019.
Per Swedish clinical routine, the ER and PR positivity cut-offs were > 10% stained nuclei. For patients with missing HER2 status, HER2 status was obtained from dual gene protein staining of HER2 on tissue microarrays, which showed 97.7% agreement with available pathological assessment [30]. Tumor-specific CAV1 staining was obtained and dichotomized, as previously described, into positive/negative for malignant cells and strong/not strong for stromal cells [13, 31]. Anthropometric measurements were dichotomized as in the previous study [13].
Genotyping
From the leukocyte portion of whole blood, DNA was extracted using DNeasy® Blood and Tissue kit and processed with QiaCube according (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. SNP genotyping was performed by the Centre for Translational Genomics at Lund University using Oncoarray by Illumina [32], specifically designed to evaluate genetic variants for association with the multiple cancers types (including breast). Details on the genotyping calling has been previously described [32]. Standard quality control was performed on all scans. All samples with low call rates (< 1 × 10–5), single-nucleotide polymorphisms (SNPs) with minor allele frequency < 1% or call rate < 99% were excluded. For CAV1 SNPs, genotype intensity cluster plots were examined manually to judge reliability [33]. Five out of six CAV1 SNPs (rs10256914, rs959173, rs3807989, rs3815412, and rs8713) passed quality control and were in Hardy–Weinberg equilibrium, while the excluded SNP had a minor allele frequency < 1%. The first four SNPs are intronic and rs8713 is a 3’ UTR variant.
CAV1 haplotype/diplotype construction
Each SNP was cross-tabulated against the other four SNPs and based on the most likely combinations, the haplotypes and diplotypes were constructed. The genotypes for rs10256914 and rs8713 were missing for one patient each and were imputed based on other genotypes (Fig. 2). The haplotypes were compared to a reference European population (1000genome project) from LDlinkR [34] (supplementary table S1). The major allele for all five SNPs were defined according to dbSNP and used as reference for all statistical analyses. Only haplotypes over 10% were analyzed and compared to no copy of each respective haplotype in the analyses. Two haplotypes (CTGTA and TTACA) were dichotomized into any (1+) and none (0) due to low frequency of homozygotes (Fig. 2).
Genomic region of CAV1 along with linkage disequilibrium heatmap and visual illustration of the linkage relationship between CAV1 SNPs. Continuous lines indicate common combinations while dotted lines indicate less likely combinations. Frequencies of CAV1 SNPs, combined genotypes, diplotypes, and haplotypes for the 1017 breast cancer patients
Database searches for proxy and putatively functional variants and expression quantitative trait loci in linkage disequilibrium with the five SNPs were performed using LDLinkR [34] in R (v4.0.2). ‘LDheatmap’ and ‘ggplot2’ were used to generate linkage disequilibrium heatmaps and forest plots, respectively.
Statistical analyses
The five individual SNPs and derived haplotypes were analyzed in relation to patient and tumor characteristics with chi-square test or linear-by-linear trend test (when appropriate) for categorical variables and Mann Whitney U-test or Kruskal–Wallis (when appropriate) for continuous variables.
Endpoints used for survival were locoregional recurrence, contralateral breast cancer, any first breast cancer event, distant metastasis, and death due to any cause. Locoregional recurrence-free interval (LRFI), contralateral breast cancer-free interval (CBCFI), breast cancer-free interval (BCFI), and distant metastasis-free interval (DMFI) were calculated from inclusion until the first event. Patients without any recurrences were censored at the time of the last follow-up before emigration, death, or last follow-up by June 30, 2019. Overall survival (OS) was defined as the time until death or last follow-up by June 30, 2019.
For survival analyses, univariable analyses were conducted with Log-rank tests and Kaplan–Meier curves. For multivariable survival analyses, Cox proportional hazards models were used. Two models were used: model 1 that was adjusted for age and tumor characteristics and model 2 that was further adjusted for adjuvant treatments. Schoenfeld’s residuals were used to test the proportional hazard assumption for the genotypes and haplotypes in model 2. Survival analyses with CBCFI as endpoint were restricted to patients without bilateral tumors. To investigate effect modifications between the CAV1 genotypes and tumor-specific CAV1 (both in malignant and stromal cells) on clinical outcome, formal two-way interactions analyses were performed in model 2. Further, since radiotherapy is mainly given to prevent locoregional disease, exploratory analysis were also performed stratified by radiotherapy for LRFI to elucidate whether the genotypes were associated with radioresistance [6, 7].
For sensitivity analyses, Fine-Gray subdistribution hazard models for two endpoints (locoregional recurrence and contralateral breast cancer) were fitted and adjusted according to multivariable model 2, to account for death and other types of breast cancer events as a competing risk. Further sensitivity analyses were conducted with additional adjustment for BMI, HER2, and tumor-specific CAV1. To accommodate for missing data for these covariates, multiple imputation by chained equations were used and the pooled results were compared to the complete case results as previously performed [13]. Since the CC genotype was more common among TNBC, an additional analysis of rs3815412 in relation to CBCFI was conducted adjusting for TNBC status.
All statistical analyses were conducted in STATA version 17.0 (StataCorp, College Station, TX, US). A P value < 0.05 was considered significant. All P values were two tailed. Nominal P values are presented without adjustment for multiple testing due to the exploratory nature of this study [35].
Results
Patient and tumor characteristics in relation CAV1 genotypes and haplotypes
Database searches revealed that all five CAV1 SNPs were linked to other genetic variants in CAV1 regulating its expression in adipocytes, in particular the rs3807989 A-allele and rs3815412 C-allele were linked genotypes associated with lower CAV1 gene expression. None of the five CAV1 SNPs were associated with patient characteristics. The TTGTA haplotype was associated with age at inclusion (P = 0.001), where patients having no haplotype were younger than other patients. No other associations between patient characteristics and haplotypes were found. Moreover, there were no associations between CAV1 SNPs and haplotypes and tumor characteristics with the exception of an association between rs959173 and nodal status (P = 0.032). Tumor-specific strong CAV1 in stromal cells and positive CAV1 in malignant cells were similar across CAV1 genotypes and haplotypes. Table 1 presents descriptive statistics for all 1017 patients as well as for SNP rs3815412 and the TTACA haplotype, which were related to prognosis.
CAV1 genotype and haplotype in relation to prognosis
The patients were followed for up to 15 years. Median follow-up for the patients still at risk (n = 734) was 9.05 years (interquartile range 7.03–11.1). There were 195 patients with any breast cancer event during follow-up (61 with locoregional recurrence, 48 with contralateral breast cancer, and 122 with distant metastasis). During follow-up, 188 patients died, of which 100 had a prior breast cancer event. The hazards for genotypes and haplotypes were proportional during follow-up.
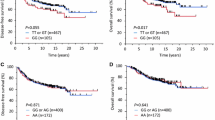
The rs3815412 CC genotype was associated with a borderline increased risk of any breast cancer event (Table 2 and supplementary figure S1) that appeared to be driven by an increased risk for contralateral breast cancer, adjusted hazard ratio (HRadj) 4.26 (95% CI 1.86–9.73; Fig. 3). There was no interaction between the rs3815412 SNP and tumor-specific CAV1 in malignant cells on CBCFI. The effect estimates became marginally higher after adjustment for TNBC status. No interaction analysis was performed because there were only three contralateral events in the TNBC subgroup. After further adjustment for BMI, HER2 status, and positive CAV1 cytoplasmic staining in malignant cells, the association remained statistically significant in both the complete case and multiple imputation models (supplementary table S2). Controlling for competing risk (other breast cancer events and death) did not substantially change the effect estimates (supplementary table S3). Furthermore, a weak association between the rs3807989 AA genotype and any breast cancer event was observed (supplementary table S4). However, the AA genotype was in complete linkage with the rs3815412 CC genotype, which appeared to drive the association. No other CAV1 SNPs were associated with clinical outcome (supplementary table S4).
Kaplan–Meier estimates of (a, c) locoregional recurrence-free interval with corresponding (b, d) forest plots of adjusted hazard ratios (95% confidence intervals), contralateral breast cancer-free interval (e, g) with corresponding (f, h) forest plots of adjusted hazard ratios (95% confidence intervals) in relation to the CAV1 rs3815412 genotype and TTACA haplotype in all patients. The number of patients is indicated at each time-point. The study is ongoing; thus, the number of patients decreases with time
Among the five common haplotypes, only TTACA was associated with outcome (Table 2 and supplementary table S5). Having at least one copy of the TTACA haplotype conferred borderline increased risk for any breast cancer event HRadj 1.39 (95% CI 0.96–2.01; Table 2 and supplementary figure S2), driven by an increased risk for locoregional recurrence HRadj 2.24 (95% CI 1.24–4.04; Fig. 1 and Table 2). The association was more pronounced in the 366 non-radiotherapy-treated patients HRadj 3.70 (95% CI 1.22–11.21) compared to the 644 radiotherapy-treated patients HRadj 1.80 (95% CI 0.77–4.23) but the effect modification was not significant (Pinteraction = 0.21). There was also no interaction between tumor-specific CAV1 in stromal cells and TTACA haplotype on LRFI. After further adjustment for BMI, HER2 status, and strong CAV1 staining in stromal cells, the association remained statistically significant in both the complete case and multiple imputation models (supplementary table S2). Controlling for competing risks did not substantially change the effect estimates (supplementary table S3).
Discussion
Both CAV1 genotypes and haplotypes were associated with risk of metachronous contralateral breast cancer and locoregional recurrence in breast cancer. The rs3815412 CC genotype was associated with a fourfold increased risk for metachronous contralateral breast cancer, and the TTACA haplotype was associated with a twofold increased risk for locoregional recurrence. We previously reported that tumor-specific CAV1 was a predictor for both contralateral breast cancer and locoregional recurrence depending on its localization [13]. The effect of CAV1 genotypes appeared to be independent of tumor characteristics including CAV1 protein expression. This indicates that host factors and tumor microenvironment may be of importance for predicting metachronous contralateral breast cancer and locoregional recurrence.
The three SNPs rs3807989, rs3815412, and rs8713, not only distinguish the TTACA haplotype from the major haplotype (TTGTA) but also capture the genomic region surrounding the last exon of the CAV1 gene, which encodes most of the functional domains [14]. None of these five Oncoarray SNPs were in coding regions but may be involved in splicing, transcription and translation of CAV1, regulating the expression of different isoforms.
Especially two of the genotyped SNPs are linked to other SNPs in the CAV1 gene that regulate CAV1 expression in adipocytes. The genotypes associated with increased risk for non-distant events in our study were associated with lower CAV1 expression in adipocytes. Loss of CAV1 in adipocytes leads to impaired internalization and storage of lipids, lipodystrophy, hypertriglyceridemia, and metabolic syndrome but notably not to increased adiposity [19, 20]. This would correlate to the metabolically obese normal-weight phenotype [36], which constitutes a unique adipose tissue microenvironment similar to obesity induced tumor microenvironment. The metabolically obese normal-weight phenotype is not well captured by BMI [36, 37]. In line with this, we found no association between BMI and CAV1 genotypes in our cohort.
The effect of the tumor microenvironment caused by the obese normal-weight phenotype on breast cancer is less well understood [36], it is possible that similar mechanisms driving breast cancer progression are at play as in the obese microenvironment [36, 37]. The knockdown of CAV1 leads to increased expression of aromatase in adipocytes [23], increasing the free estrogen in the surrounding tissues promoting breast cancer tumorigenesis [36, 37]. Also, CAV1 deficiency leads to inability to properly stabilize the insulin receptor, rendering the adipocytes unresponsive to insulin [38] and causing inflammation [39]. This tumor promoting inflammation might be mediated by M2 macrophages that promote tumorigenesis [22] and are linked to CAV1 expression in adipocytes [21]. Taken together, this indicates that the obese normal-weight phenotype, which might be captured by the CAV1 genotype, favors the development of metachronous contralateral breast cancer and locoregional recurrence whereas obesity favors distant recurrences. To summarize, decreased CAV1, which the CAV1 SNPs were related to, leads to several changes resulting in an unfavorable adipose tissue microenvironment [36] that may promote recurrences in especially in breast tissue, which would explain our findings. The impact of CAV1 TTACA haplotype on locoregional recurrence risk was less pronounced in radiotherapy-treated patients compared to non-treated patients. The finding merits further investigation to elucidate whether radiotherapy to prevent locoregional recurrences might be especially beneficial for patients with the CAV1 TTACA haplotype. CAV1 expression in tumors has been linked to radioresistance in several cancers [18]. The relationship between CAV1 genotypes and radioresistance is still unknown. Further studies are needed.
The strengths of this study includes, a population-based patient cohort considered representative for its catchment area with reliable clinicopathological and anthropometric data [40]. The most common reason for not participating was the lack of available research nurses. Further approximately 5% of patients had an unclear diagnosis at the time of surgery and were therefore not included at the preoperative visit. Previous studies demonstrated that participants of the BCblood cohort were similar to all operated patients with regards to age and hormone receptor status [40, 41]. Additionally, tumor-specific CAV1 data were available [13], enabling a unique dataset with long-term follow-up for analysis. CAV1 genotyping was done using a SNParray designed to investigate genetic variations in relation to cancer [32]. Nonetheless, it would valuable to investigate in-depth the CAV1 genomic region to elucidate causal relationships between CAV1 genotypes, adipocytes, and breast cancer.
Most cases of metachronous contralateral breast cancer are considered to be new primary cancer [11]. This would imply that the rs3815412 CC genotype might be a risk factor for primary breast cancer. To our knowledge, genome-wide association studies did not find associations between CAV1 polymorphisms or the genomic region where it is located and breast cancer risk [42, 43]. However, in case–control and cohort studies, the rs3807987 SNP, which is in linkage with the rs3815412, was associated with breast cancer risk Asian and Middle Eastern populations [27]. Further, several SNPs in multiple genes are more strongly associated with either ER-positive or negative disease [42, 44]. In our cohort, there were very few metachronous contralateral breast cancers in the ER-negative subgroup, making subgroup analyses meaningless. To confirm our findings, large and well-designed studies in various populations are needed.
Metachronous contralateral breast cancer and locoregional recurrence have few established specific prognostic markers, yet impact outcome in breast cancer [8,9,10]. For locoregional recurrence, prognostic factors related to tumor phenotype have been proposed [45,46,47]. Specific prognostic factors for metachronous contralateral breast cancer [7] are mostly related to established factors for breast cancer risk. Beyond existing tumor-related prognostic factors, CAV1 genotypes might offer new prognostic information related to the host.
In conclusion, CAV1 polymorphisms were shown to be associated with an increased risk for contralateral breast cancer and locoregional recurrence. If confirmed, the findings may identify patients that could derive benefit from more tailored treatment to prevent non-distant breast cancer events.
Data availability
Clinical data are not publicly available due to privacy laws. Questions regarding data can be directed to the corresponding author.
Abbreviations
- BCFI:
-
Breast cancer-free interval
- BMI:
-
Body mass index
- CAV1:
-
Caveolin-1
- CBCFI:
-
Contralateral breast cancer-free interval
- DMFI:
-
Distant metastasis-free interval
- EDTA:
-
Ethylenediaminetetraacetic acid
- ER:
-
Estrogen receptor
- HER2:
-
Human epidermal growth factor receptor 2
- HR:
-
Hazard ratio
- IHC:
-
Immunohistochemistry
- LRFI:
-
Locoregional recurrence-free interval
- NoE:
-
Number of events
- OS:
-
Overall survival
- SNP:
-
Single-nucleotide polymorphism
- PR:
-
Progesterone receptor
- TGFβ:
-
Transforming growth factor-beta
- TNBC:
-
Triple-negative breast cancer
References
Harbeck N, Penault-Llorca F, Cortes J, Gnant M, Houssami N, Poortmans P, Ruddy K, Tsang J, Cardoso F (2019) Breast cancer Nat Rev Dis Primers 5:66. https://doi.org/10.1038/s41572-019-0111-2
Harris LN, Ismaila N, McShane LM, Andre F, Collyar DE, Gonzalez-Angulo AM, Hammond EH, Kuderer NM, Liu MC, Mennel RG, Van Poznak C, Bast RC, Hayes DF, American Society of Clinical O (2016) Use of biomarkers to guide decisions on adjuvant systemic therapy for women with early-stage invasive breast cancer: American Society of Clinical Oncology Clinical Practice Guideline. J Clin Oncol 34:1134–1150. https://doi.org/10.1200/JCO.2015.65.2289
Kwa M, Makris A, Esteva FJ (2017) Clinical utility of gene-expression signatures in early stage breast cancer. Nat Rev Clin Oncol 14:595–610. https://doi.org/10.1038/nrclinonc.2017.74
Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Lønning PE, Børresen-Dale AL (2001) Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA 98:10869–10874. https://doi.org/10.1073/pnas.191367098
Gnant M, Filipits M, Greil R, Stoeger H, Rudas M, Bago-Horvath Z, Mlineritsch B, Kwasny W, Knauer M, Singer C, Jakesz R, Dubsky P, Fitzal F, Bartsch R, Steger G, Balic M, Ressler S, Cowens JW, Storhoff J, Ferree S, Schaper C, Liu S, Fesl C, Nielsen TO (2014) Predicting distant recurrence in receptor-positive breast cancer patients with limited clinicopathological risk: using the PAM50 Risk of Recurrence score in 1478 postmenopausal patients of the ABCSG-8 trial treated with adjuvant endocrine therapy alone. Ann Oncol 25:339–345. https://doi.org/10.1093/annonc/mdt494
Lowery AJ, Kell MR, Glynn RW, Kerin MJ, Sweeney KJ (2012) Locoregional recurrence after breast cancer surgery: a systematic review by receptor phenotype. Breast Cancer Res Treat 133:831–841. https://doi.org/10.1007/s10549-011-1891-6
Akdeniz D, Schmidt MK, Seynaeve CM, McCool D, Giardiello D, van den Broek AJ, Hauptmann M, Steyerberg EW, Hooning MJ (2019) Risk factors for metachronous contralateral breast cancer: a systematic review and meta-analysis. Breast 44:1–14. https://doi.org/10.1016/j.breast.2018.11.005
Bantema-Joppe EJ, van den Heuvel ER, de Munck L, de Bock GH, Smit WG, Timmer PR, Dolsma WV, Jansen L, Schröder CP, Siesling S, Langendijk JA, Maduro JH (2013) Impact of primary local treatment on the development of distant metastases or death through locoregional recurrence in young breast cancer patients. Breast Cancer Res Treat 140:577–585. https://doi.org/10.1007/s10549-013-2650-7
de Bock GH, Putter H, Bonnema J, van der Hage JA, Bartelink H, van de Velde CJ (2009) The impact of loco-regional recurrences on metastatic progression in early-stage breast cancer: a multistate model. Breast Cancer Res Treat 117:401–408. https://doi.org/10.1007/s10549-008-0300-2
Geurts YM, Witteveen A, Bretveld R, Poortmans PM, Sonke GS, Strobbe LJA, Siesling S (2017) Patterns and predictors of first and subsequent recurrence in women with early breast cancer. Breast Cancer Res Treat 165:709–720. https://doi.org/10.1007/s10549-017-4340-3
Begg CB, Ostrovnaya I, Geyer FC, Papanastasiou AD, Ng CKY, Sakr RA, Bernstein JL, Burke KA, King TA, Piscuoglio S, Mauguen A, Orlow I, Weigelt B, Seshan VE, Morrow M, Reis-Filho JS (2018) Contralateral breast cancers: independent cancers or metastases? Int J Cancer 142:347–356. https://doi.org/10.1002/ijc.31051
Alkner S, Tang MH, Brueffer C, Dahlgren M, Chen Y, Olsson E, Winter C, Baker S, Ehinger A, Rydén L, Saal LH, Fernö M, Gruvberger-Saal SK (2015) Contralateral breast cancer can represent a metastatic spread of the first primary tumor: determination of clonal relationship between contralateral breast cancers using next-generation whole genome sequencing. Breast Cancer Res 17:102. https://doi.org/10.1186/s13058-015-0608-x
Godina C, Indira-Chandran V, Barbachowska M, Tryggvadottir H, Nodin B, Visse E, Borgquist S, Jirström K, Isaksson K, Bosch A, Belting M, Jernström H (2022) Interplay between Caveolin-1 and body and tumor size affects clinical outcomes in breast cancer. Transl Oncol 22:101464. https://doi.org/10.1016/j.tranon.2022.101464
Engelman JA, Zhang XL, Galbiati F, Lisanti MP (1998) Chromosomal localization, genomic organization, and developmental expression of the murine caveolin gene family (Cav-1, -2, and -3). Cav-1 and Cav-2 genes map to a known tumor suppressor locus (6–A2/7q31). FEBS Lett 429:330–336. https://doi.org/10.1016/s0014-5793(98)00619-x
Patani N, Martin LA, Reis-Filho JS, Dowsett M (2012) The role of caveolin-1 in human breast cancer. Breast Cancer Res Treat 131:1–15. https://doi.org/10.1007/s10549-011-1751-4
Simón L, Campos A, Leyton L, Quest AFG (2020) Caveolin-1 function at the plasma membrane and in intracellular compartments in cancer. Cancer Metastas Rev 39:435–453. https://doi.org/10.1007/s10555-020-09890-x
Williams TM, Lisanti MP (2004) The caveolin proteins. Genome Biol 5:214. https://doi.org/10.1186/gb-2004-5-3-214
Ketteler J, Klein D (2018) Caveolin-1, cancer and therapy resistance. Int J Cancer 143:2092–2104. https://doi.org/10.1002/ijc.31369
Pilch PF, Liu L (2011) Fat caves: caveolae, lipid trafficking and lipid metabolism in adipocytes. Trends Endocrinol Metab 22:318–324. https://doi.org/10.1016/j.tem.2011.04.001
Razani B, Combs TP, Wang XB, Frank PG, Park DS, Russell RG, Li M, Tang B, Jelicks LA, Scherer PE, Lisanti MP (2002) Caveolin-1-deficient mice are lean, resistant to diet-induced obesity, and show hypertriglyceridemia with adipocyte abnormalities. J Biol Chem 277:8635–8647. https://doi.org/10.1074/jbc.M110970200
Briand N, Le Lay S, Sessa WC, Ferré P, Dugail I (2011) Distinct roles of endothelial and adipocyte caveolin-1 in macrophage infiltration and adipose tissue metabolic activity. Diabetes 60:448–453. https://doi.org/10.2337/db10-0856
Liu J, Geng X, Hou J, Wu G (2021) New insights into M1/M2 macrophages: key modulators in cancer progression. Cancer Cell Int 21:389. https://doi.org/10.1186/s12935-021-02089-2
Mukherjee R, Kim SW, Choi MS, Yun JW (2014) Sex-dependent expression of caveolin 1 in response to sex steroid hormones is closely associated with development of obesity in rats. PLoS ONE 9:90918. https://doi.org/10.1371/journal.pone.0090918
Wang YY, Lehuédé C, Laurent V, Dirat B, Dauvillier S, Bochet L, Le Gonidec S, Escourrou G, Valet P, Muller C (2012) Adipose tissue and breast epithelial cells: a dangerous dynamic duo in breast cancer. Cancer Lett 324:142–151. https://doi.org/10.1016/j.canlet.2012.05.019
García-Estévez L, Cortés J, Pérez S, Calvo I, Gallegos I, Moreno-Bueno G (2021) Obesity and breast cancer: a paradoxical and controversial relationship influenced by menopausal status. Front Oncol 11:705911. https://doi.org/10.3389/fonc.2021.705911
Aali Y, Shiraseb F, Abaj F, Koohdani F, Mirzaei K (2021) The interactions between dietary fats intake and Caveolin 1 rs 3807992 polymorphism with fat distribution in overweight and obese women: a cross-sectional study. BMC Med Genomics 14:265. https://doi.org/10.1186/s12920-021-01114-7
Yan C, Sun C, Ding X, Rizeq FK, Ren M, Yang F, Chen Y, Wang B (2019) Association of CAV1 polymorphisms with the risks of breast cancer: a systematic review and meta-analysis. Pathol Res Pract 215:152518. https://doi.org/10.1016/j.prp.2019.152518
Chen P, Zhang YL, Xue B, Wang JR (2021) CAV1 rs7804372 (T29107A) polymorphism might be a potential risk for digestive cancers: a protocol for systematic review and meta analysis. Medicine (Baltimore) 100:e26186. https://doi.org/10.1097/md.0000000000026186
Fan S, Meng J, Zhang L, Zhang X, Liang C (2019) CAV1 polymorphisms rs1049334, rs1049337, rs7804372 might be the potential risk in tumorigenicity of urinary cancer: a systematic review and meta-analysis. Pathol Res Pract 215:151–158. https://doi.org/10.1016/j.prp.2018.11.009
Sandén E, Khazaei S, Tryggvadottir H, Borgquist S, Isaksson K, Jirström K, Jernström H (2020) Re-evaluation of HER2 status in 606 breast cancers-gene protein assay on tissue microarrays versus routine pathological assessment. Virchows Arch. https://doi.org/10.1007/s00428-020-02768-x
Indira Chandran V, Månsson AS, Barbachowska M, Cerezo-Magaña M, Nodin B, Joshi B, Koppada N, Saad OM, Gluz O, Isaksson K, Borgquist S, Jirström K, Nabi IR, Jernström H, Belting M (2020) Hypoxia attenuates trastuzumab uptake and trastuzumab-emtansine (T-DM1) cytotoxicity through redistribution of phosphorylated caveolin-1. Mol Cancer Res 18:644–656. https://doi.org/10.1158/1541-7786.Mcr-19-0856
Amos CI, Dennis J, Wang Z, Byun J, Schumacher FR, Gayther SA, Casey G, Hunter DJ, Sellers TA, Gruber SB, Dunning AM, Michailidou K, Fachal L, Doheny K, Spurdle AB, Li Y, Xiao X, Romm J, Pugh E, Coetzee GA, Hazelett DJ, Bojesen SE, Caga-Anan C, Haiman CA, Kamal A, Luccarini C, Tessier D, Vincent D, Bacot F, Van Den Berg DJ, Nelson S, Demetriades S, Goldgar DE, Couch FJ, Forman JL, Giles GG, Conti DV, Bickeböller H, Risch A, Waldenberger M, Brüske-Hohlfeld I, Hicks BD, Ling H, McGuffog L, Lee A, Kuchenbaecker K, Soucy P, Manz J, Cunningham JM, Butterbach K, Kote-Jarai Z, Kraft P, FitzGerald L, Lindström S, Adams M, McKay JD, Phelan CM, Benlloch S, Kelemen LE, Brennan P, Riggan M, O’Mara TA, Shen H, Shi Y, Thompson DJ, Goodman MT, Nielsen SF, Berchuck A, Laboissiere S, Schmit SL, Shelford T, Edlund CK, Taylor JA, Field JK, Park SK, Offit K, Thomassen M, Schmutzler R, Ottini L, Hung RJ, Marchini J, Amin Al Olama A, Peters U, Eeles RA, Seldin MF, Gillanders E, Seminara D, Antoniou AC, Pharoah PD, Chenevix-Trench G, Chanock SJ, Simard J, Easton DF (2017) The oncoarray consortium: a network for understanding the genetic architecture of common cancers. Cancer Epidemiol Biomark Prev 26:126–135. https://doi.org/10.1158/1055-9965.Epi-16-0106
Michailidou K, Hall P, Gonzalez-Neira A, Ghoussaini M, Dennis J, Milne RL, Schmidt MK, Chang-Claude J, Bojesen SE, Bolla MK, Wang Q, Dicks E, Lee A, Turnbull C, Rahman N, Fletcher O, Peto J, Gibson L, Dos Santos SI, Nevanlinna H, Muranen TA, Aittomäki K, Blomqvist C, Czene K, Irwanto A, Liu J, Waisfisz Q, Meijers-Heijboer H, Adank M, van der Luijt RB, Hein R, Dahmen N, Beckman L, Meindl A, Schmutzler RK, Müller-Myhsok B, Lichtner P, Hopper JL, Southey MC, Makalic E, Schmidt DF, Uitterlinden AG, Hofman A, Hunter DJ, Chanock SJ, Vincent D, Bacot F, Tessier DC, Canisius S, Wessels LF, Haiman CA, Shah M, Luben R, Brown J, Luccarini C, Schoof N, Humphreys K, Li J, Nordestgaard BG, Nielsen SF, Flyger H, Couch FJ, Wang X, Vachon C, Stevens KN, Lambrechts D, Moisse M, Paridaens R, Christiaens MR, Rudolph A, Nickels S, Flesch-Janys D, Johnson N, Aitken Z, Aaltonen K, Heikkinen T, Broeks A, Veer LJ, van der Schoot CE, Guénel P, Truong T, Laurent-Puig P, Menegaux F, Marme F, Schneeweiss A, Sohn C, Burwinkel B, Zamora MP, Perez JI, Pita G, Alonso MR, Cox A, Brock IW, Cross SS, Reed MW, Sawyer EJ, Tomlinson I, Kerin MJ, Miller N, Henderson BE et al (2013) Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet 45:353–361. https://doi.org/10.1038/ng.2563
Myers TA, Chanock SJ, Machiela MJ (2020) LDlinkR: an R package for rapidly calculating linkage disequilibrium statistics in diverse populations. Front Genet 11:157. https://doi.org/10.3389/fgene.2020.00157
Victor A, Elsässer A, Hommel G, Blettner M (2010) Judging a plethora of p-values: how to contend with the problem of multiple testing–part 10 of a series on evaluation of scientific publications. Dtsch Arztebl Int 107:50–56. https://doi.org/10.3238/arztebl.2010.0050
Quail DF, Dannenberg AJ (2019) The obese adipose tissue microenvironment in cancer development and progression. Nat Rev Endocrinol 15:139–154. https://doi.org/10.1038/s41574-018-0126-x
Mubtasim N, Moustaid-Moussa N, Gollahon L (2022) The complex biology of the obesity-induced, metastasis-promoting tumor microenvironment in breast cancer. Int J Mol Sci. https://doi.org/10.3390/ijms23052480
Cohen AW, Razani B, Wang XB, Combs TP, Williams TM, Scherer PE, Lisanti MP (2003) Caveolin-1-deficient mice show insulin resistance and defective insulin receptor protein expression in adipose tissue. Am J Physiol Cell Physiol 285:C222-235. https://doi.org/10.1152/ajpcell.00006.2003
Shimobayashi M, Albert V, Woelnerhanssen B, Frei IC, Weissenberger D, Meyer-Gerspach AC, Clement N, Moes S, Colombi M, Meier JA, Swierczynska MM, Jenö P, Beglinger C, Peterli R, Hall MN (2018) Insulin resistance causes inflammation in adipose tissue. J Clin Invest 128:1538–1550. https://doi.org/10.1172/jci96139
Persson M, Simonsson M, Markkula A, Rose C, Ingvar C, Jernström H (2016) Impacts of smoking on endocrine treatment response in a prospective breast cancer cohort. Br J Cancer 115:382–390. https://doi.org/10.1038/bjc.2016.174
Lundin KB, Henningson M, Hietala M, Ingvar C, Rose C, Jernstrom H (2011) Androgen receptor genotypes predict response to endocrine treatment in breast cancer patients. Br J Cancer 105:1676–1683. https://doi.org/10.1038/bjc.2011.441
Lilyquist J, Ruddy KJ, Vachon CM, Couch FJ (2018) Common genetic variation and breast cancer risk-past, present, and future. Cancer Epidemiol Biomark Prev 27:380–394. https://doi.org/10.1158/1055-9965.Epi-17-1144
Michailidou K, Lindström S, Dennis J, Beesley J, Hui S, Kar S, Lemaçon A, Soucy P, Glubb D, Rostamianfar A, Bolla MK, Wang Q, Tyrer J, Dicks E, Lee A, Wang Z, Allen J, Keeman R, Eilber U, French JD, Qing Chen X, Fachal L, McCue K, McCart Reed AE, Ghoussaini M, Carroll JS, Jiang X, Finucane H, Adams M, Adank MA, Ahsan H, Aittomäki K, Anton-Culver H, Antonenkova NN, Arndt V, Aronson KJ, Arun B, Auer PL, Bacot F, Barrdahl M, Baynes C, Beckmann MW, Behrens S, Benitez J, Bermisheva M, Bernstein L, Blomqvist C, Bogdanova NV, Bojesen SE, Bonanni B, Børresen-Dale AL, Brand JS, Brauch H, Brennan P, Brenner H, Brinton L, Broberg P, Brock IW, Broeks A, Brooks-Wilson A, Brucker SY, Brüning T, Burwinkel B, Butterbach K, Cai Q, Cai H, Caldés T, Canzian F, Carracedo A, Carter BD, Castelao JE, Chan TL, David Cheng TY, Seng Chia K, Choi JY, Christiansen H, Clarke CL, Collée M, Conroy DM, Cordina-Duverger E, Cornelissen S, Cox DG, Cox A, Cross SS, Cunningham JM, Czene K, Daly MB, Devilee P, Doheny KF, Dörk T, Dos-Santos-Silva I, Dumont M, Durcan L, Dwek M, Eccles DM, Ekici AB, Eliassen AH, Ellberg C, Elvira M, Engel C et al (2017) Association analysis identifies 65 new breast cancer risk loci. Nature 551:92–94. https://doi.org/10.1038/nature24284
Milne RL, Kuchenbaecker KB, Michailidou K, Beesley J, Kar S, Lindström S, Hui S, Lemaçon A, Soucy P, Dennis J, Jiang X, Rostamianfar A, Finucane H, Bolla MK, McGuffog L, Wang Q, Aalfs CM, Adams M, Adlard J, Agata S, Ahmed S, Ahsan H, Aittomäki K, Al-Ejeh F, Allen J, Ambrosone CB, Amos CI, Andrulis IL, Anton-Culver H, Antonenkova NN, Arndt V, Arnold N, Aronson KJ, Auber B, Auer PL, Ausems M, Azzollini J, Bacot F, Balmaña J, Barile M, Barjhoux L, Barkardottir RB, Barrdahl M, Barnes D, Barrowdale D, Baynes C, Beckmann MW, Benitez J, Bermisheva M, Bernstein L, Bignon YJ, Blazer KR, Blok MJ, Blomqvist C, Blot W, Bobolis K, Boeckx B, Bogdanova NV, Bojesen A, Bojesen SE, Bonanni B, Børresen-Dale AL, Bozsik A, Bradbury AR, Brand JS, Brauch H, Brenner H, Bressac-de Paillerets B, Brewer C, Brinton L, Broberg P, Brooks-Wilson A, Brunet J, Brüning T, Burwinkel B, Buys SS, Byun J, Cai Q, Caldés T, Caligo MA, Campbell I, Canzian F, Caron O, Carracedo A, Carter BD, Castelao JE, Castera L, Caux-Moncoutier V, Chan SB, Chang-Claude J, Chanock SJ, Chen X, Cheng TD, Chiquette J, Christiansen H, Claes KBM, Clarke CL, Conner T, Conroy DM, Cook J et al (2017) Identification of ten variants associated with risk of estrogen-receptor-negative breast cancer. Nat Genet 49:1767–1778. https://doi.org/10.1038/ng.3785
Davey MG, Cleere EF, O’Donnell JP, Gaisor S, Lowery AJ, Kerin MJ (2022) Value of the 21-gene expression assay in predicting locoregional recurrence rates in estrogen receptor-positive breast cancer: a systematic review and network meta-analysis. Breast Cancer Res Treat 193:535–544. https://doi.org/10.1007/s10549-022-06580-w
Narayan P, Flynn J, Zhang Z, Gillespie EF, Mueller B, Xu AJ, Cuaron J, McCormick B, Khan AJ, Cahlon O, Powell SN, Wen H, Braunstein LZ (2021) Perineural invasion as a risk factor for locoregional recurrence of invasive breast cancer. Sci Rep 11:12781. https://doi.org/10.1038/s41598-021-92343-4
Van der Vorst A, Kindts I, Laenen A, Neven P, Janssen H, Weltens C (2022) Validation of a prognostic scoring system for postmastectomy locoregional recurrence in breast cancer. Breast 64:29–34. https://doi.org/10.1016/j.breast.2022.04.007
Acknowledgements
We would like to thank our research nurses Linda Ågren, Helén Thell, Jessica Åkesson, Anette Ahlin Gullers, Monika Eberhard Mészaros, Maj-Britt Hedenblad, Karin Henriksson, and Anette Möller. Additionally, we would like to thank Erika Bågeman, Maria Henningson, Maria Hjertberg, Maria Ygland Rödström, and Andrea Markkula for data entry. The authors thank Helén Thell and Björn Nodin for help with DNA extraction and processing of the samples. The authors would like to Acknowledge Clinical Genomics Lund, SciLifeLab and Center for Translational Genomics (CTG), Lund University, for providing expertise and service with sequencing and analysis.
Funding
Open access funding provided by Lund University. The Swedish Cancer Society (CAN 20 0763), the Faculty of Medicine at Lund University, the Mrs Berta Kamprad Foundation, the South Swedish Health Care Region (Region Skåne ALF 40620), and the Skåne University Hospital fund. AB holds a young researcher award from ALF (Region Skåne). HT was funded by Region Skåne ST-ALF. The funders had no role in study design and conduct of the study, data collection and analysis, data interpretation, or manuscript preparation and decision to submit the manuscript for publication.
Author information
Authors and Affiliations
Contributions
CG, HJ: conceptualization, CG, HJ, HT: Data curation, CG, HJ: Formal Analysis, HJ, KI: Funding acquisition, HJ: Investigation, CG, HJ: Methodology, HJ: Project administration, HJ, KJ, KI, MB, SB, HT: Resources, HJ: Supervision, CG: Visualization, CG, HJ: Writing—original draft All authors: Writing—review & editing.
Corresponding author
Ethics declarations
Conflict of interest
Ana Bosch has received institutional honoraria from Pfizer, Roche, and Lilly for consultation and lectures. She has participated in Advisory Board meetings for Pfizer and Novartis. Co-founder and chair of the board for SACRA therapeutics. The other authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
10549_2023_6919_MOESM1_ESM.pdf
Kaplan-Meier estimates of (A) breast cancer-free interval with corresponding (B) forest plot of adjusted hazard ratios (95% confidence intervals), (C) distant metastasis-free interval with corresponding (D) forest plot of adjusted hazard ratios (95% confidence intervals), (E) overall survival with corresponding (F) forest plot of adjusted hazard ratios (95% confidence intervals) in relation to CAV1 rs3815412 genotype in all patients. The number of patients is indicated at each time-point. The study is ongoing; thus, the number of patients decreases with time. Supplementary file1 (PDF 356 kb)
10549_2023_6919_MOESM2_ESM.pdf
Kaplan-Meier estimates of (A) breast cancer-free interval with corresponding (B) forest plot of adjusted hazard ratios (95% confidence intervals), (C) distant metastasis-free interval with corresponding (D) forest plot of adjusted hazard ratios (95% confidence intervals), (E) overall survival with corresponding (F) forest plot of adjusted hazard ratios (95% confidence intervals) in relation to CAV1 TTACA haplotype in all patients. The number of patients is indicated at each time-point. The study is ongoing; thus, the number of patients decreases with time. Supplementary file2 (PDF 317 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Godina, C., Tryggvadottir, H., Bosch, A. et al. Caveolin-1 genotypes as predictor for locoregional recurrence and contralateral disease in breast cancer. Breast Cancer Res Treat 199, 335–347 (2023). https://doi.org/10.1007/s10549-023-06919-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-023-06919-x