Abstract
Purpose
There is growing interest in the application of circulating tumour DNA (ctDNA) as a sensitive tool for monitoring tumour evolution and guiding targeted therapy in patients with cancer. However, robust comparisons of different platform technologies are still required. Here we compared the InVisionSeq™ ctDNA Assay with the Oncomine™ Breast cfDNA Assay to assess their concordance and feasibility for the detection of mutations in plasma at low (< 0.5%) variant allele fraction (VAF).
Methods
Ninety-six plasma samples from 50 patients with estrogen receptor (ER)-positive metastatic breast cancer (mBC) were profiled using the InVision Assay. Results were compared to the Oncomine assay in 30 samples from 26 patients, where there was sufficient material and variants were covered by both assays. Longitudinal samples were analysed for 8 patients with endocrine resistance.
Results
We detected alterations in 59/96 samples from 34/50 patients analysed with the InVision assay, most frequently affecting ESR1, PIK3CA and TP53. Complete or partial concordance was found in 28/30 samples analysed by both assays, and VAF values were highly correlated. Excellent concordance was found for most genes, and most discordant calls occurred at VAF < 1%. In longitudinal samples from progressing patients with endocrine resistance, we detected consistent alterations in sequential samples, most commonly in ESR1 and PIK3CA.
Conclusion
This study shows that both ultra-deep next-generation sequencing (NGS) technologies can detect genomic alternations even at low VAFs in plasma samples of mBC patients. The strong agreement of the technologies indicates sufficient reproducibility for clinical use as prognosic and predictive biomarker.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Over the past few decades, therapies have been developed for the treatment of the most predominant (estrogen receptor (ER)-positive) subset of breast cancers (BC) and anti-estrogens (e.g. tamoxifen, fulvestrant) and aromatase inhibitors (e.g. letrozole, anastrozole, exemestane) are in widespread use as adjuvant and metastatic therapies [1].
However, many patients become resistant to treatment and research into the mechanisms of resistance has been hampered by the invasive approach of obtaining tissue biopsies from patients with endocrine-treatment-resistant metastatic disease (mBC). More recently, characterisation of mBC has been made more feasible by molecular profiling of circulating cell-free DNA derived from tumour cells [termed circulating tumour DNA (ctDNA)], through liquid biopsy [2, 3]. This ctDNA is released by dying cancer cells and active secretion and is considered to be more reflective of multiple metastatic subclones than tissue biopsy. Depending on the size, stage and activity of the cancer, tumour-specific alterations can be present in plasma samples in a background of cell-free DNA derived from healthy normal cells, with a variant allele fractions (VAFs) below 0.5% [4]. Therefore, there is a clinical need for the development, optimisation and validation of highly sensitive methods for detection of ctDNA.
To determine the optimum ctDNA-based approaches for guiding clinical treatment decisions, there is a need for inter-laboratory comparison of technologies used for ctDNA variant detection. There are multiple assays for ctDNA analysis, including those targeting a single/small number of variants, such as digital droplet-PCR (ddPCR) and bead-based digital PCR in emulsion (BEAMing), and broader targeted gene/mutation panels typically analysed by next-generation sequencing (NGS) [5]. To date, few comparisons have been published comparing different platforms for ctDNA measurement [6], most of which compare digital PCR (dPCR) with NGS platforms [7, 8].
This study focusses on the comparison of two different commercial NGS methods of ctDNA detection/quantification, the InVisionSeq™ ctDNA Assay, developed by Inivata Limited and the Oncomine™ Breast cfDNA Assay_v1 by Thermo Fisher Scientific. The InVision assay identifies single nucleotide variants (SNVs), insertions and deletions (INDELs) and copy number variants (CNVs) across whole genes and hotspots in a 36-gene panel [9]. Together with analysis of gene fusion events in plasma, it forms part of the InVisionFirst®-Lung comprehensive genomic profiling liquid biopsy assay that is in clinical use. The technology has demonstrated an excellent limit of detection (LOD) in analytical validation studies [9, 10], with 99.48% sensitivity for SNVs present at VAF range 0.25–0.33% and 56.25% sensitivity at VAF range 0.06–0.08% while retaining high specificity (99.9997% per base). High concordance with dPCR in patients with NSCLC was also demonstrated [10, 11]. The Oncomine™ Breast cfDNA Assay_v1 is a more focussed NGS assay which combines Ion AmpliSeq™ and Tag Sequencing technologies enabling detection of primary driver and resistance mutations (SNVs/INDELs) from ctDNA down to a level of 0.1% with ~ 81% sensitivity and 99.9% specificity [12].
In this report, we describe our direct comparison of both methods and show excellent correlation between both variants and VAFs detected by the two approaches. Further, we present results of targeted sequencing of ctDNA in a small group of patients with mBC, who had become resistant to endocrine therapy. We show excellent concordance between the two assays, in agreement with a recent study comparing 2 NGS platforms that found no significant differences in detection of alterations in plasma of patients with mBC [13]. These findings contrast with 2 previous studies, one that found low concordance when comparing plasma of prostate cancer patients in 2 platforms [14] and one finding substantial variability among the 4 ctDNA assays compared, with most discordance observed at VAF < 1% [15].
Patients and methods
Ethics statement
The study protocol was approved by the Riverside Research Ethics Committee (Imperial College Healthcare NHS Trust; REC reference number: 07/Q0401/20). Blood sample collection was conducted in accordance with the Declaration of Helsinki. All patients gave written informed consent before participation.
Blood processing and extraction of total cfDNA
A total of 96 blood samples from 50 unselected patients with radiologically-confirmed mBC were collected into EDTA-containing tubes (BD Biosciences) and processed to plasma within 2 h of collection. Total cfDNA was extracted from 3 mL of plasma using the QIAamp Circulating Nucleic Acids Kit (Qiagen) and quantification of eluted cfDNA was performed using the Qubit® 2.0 dsDNA high sensitivity assay (Thermo Fisher Scientific) as described previously [16].
Targeted deep sequencing
A minimum of 20 ng total cfDNA isolated from 35 plasma samples were sequenced using the two established technologies; the InVisionSeq™ ctDNA Assay (Inivata, Cambridge) v1.4 or v1.5 (Supplementary Tables S1, S2) and the Oncomine™ Breast cfDNA Assay_v1 (Supplementary Table S3) (Thermo Fisher Scientific, Waltham, MA). Due to limiting cfDNA, the remaining 61 plasma samples were analysed using the InVision assay only.
The InVision assay profiles a combination of SNVs and INDELs across 35 cancer genes (v1.4) or 36 genes (v1.5), as well as CNVs in 4 genes. The Oncomine assay analyses SNVs/INDELs covering > 150 hotspots in 10 frequently mutated genes in BC. Seven genes are overlapping and hotspot mutations present in both panels were compared (Supplementary Table S4). A more detailed explanation of sequencing analysis is provided in the Supplementary Methods.
Statistical analysis
For each sample pair the VAF and number of detected alterations (NDA) were compared between the two platforms using the Mann–Whitney U test and the Wilcoxon matched-pairs signed-ranks test. Spearman’s rank correlation coefficient and linear regression were used to determine whether the variables were correlated. Agreement in terms of detected altered genes were explored through Cohen's kappa coefficient. Statistical analysis was performed using Prism GraphPad_v6, San Diego, CA).
Analysis of sequential patient samples
Eight patients with ER-positive BC and proven distant metastases were included as a small sub-study, where samples were collected at the beginning, and/or at some point during, endocrine therapy for mBC, and again at the end of therapy when they were no longer responding. All 8 patients had received one or more additional lines of endocrine therapy prior to that undergone at the time of blood sampling. Total cfDNA levels, circulating tumour cells (CTCs) counts and protein markers serum cancer antigen 15–3 (CA15-3) and alkaline phosphatase (ALK-PHOS) were also measured (Supplementary Methods).
Results
InVisionSeq™ ctDNA assay
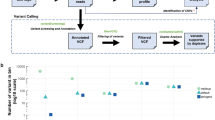
Ninety-six cfDNA samples from 50 patients were analysed with the InVisionSeq™ assay, of which 59 (61.5%) had 1 or more variants detected (Supplementary Table S5, Fig. 1). A total of 174 alterations in 20 genes were detected, 11 of which were amplifications in FGFR1 (n = 9), ERBB2 (n = 1) and MET (n = 1). SNVs comprised mainly missense but also truncating, splice-site and synonymous variants while INDELS comprised a mix of inframe and frameshift variants. VAFs ranged between 0.07 and 59.6%, with a median of 1.18%. Of 163 variants detected, 78 (47.9%) were detected at VAF ≤ 1%, and 25 (15.3%) were detected at VAF ≤ 0.25%. The most mutated genes were ESR1 (n = 53), TP53 (n = 32), PIK3CA (n = 31) and ERBB2 (n = 10). As for occurrence of mutations in each patient, 34/50 patients (68%) had at least one mutation detected, with PIK3CA [19/50 (38%)], ESR1 [17/50 (34%)] and TP53 [16/50 (32%)] being the most frequent, followed by ERBB2 (14%), FGFR1 (12%) and GATA3 (10%).
High concordance between ctDNA variants and VAF detected using the InVisionSeq™ ctDNA assay and oncomine™ breast cfDNA assay
Overlapping variants in 7 genes (AKT1, EGFR, ERBB2, ESR1, KRAS, PIK3CA and TP53) covered by both Oncomine and InVision_v1.4 assays were compared in 30 of the 35 (85.7%) samples analysed with both assays (Table 1). Five plasma samples were excluded from analysis as all alterations detected were unique to the InVision assay. The results show high concordance between the 2 assays. 47 variants were identified by at least one platform; all 31 variants detected by the InVision assay were cofirmed by Oncomine, while 16 variants in 8 samples (VAF in the range 0.05–0.46%) affecting TP53, PIK3CA and ESR1 were called only by Oncomine. Ten of 30 plasma samples (33.3%) had no variants detected by either assay. Twelve of 30 plasma samples (40%) had complete concordance with 1 or more alterations detected and 6/30 samples (20%) had partial concordance with 1 or more overlapping variants. Two of 30 samples (6.7%) showed no concordance that had variants detected by Oncomine only. Both were identified as below the minimum input DNA and one had low read depth during sequencing and were called as indeterminate for InVision_v1.4 (Fig. 2a, Table 1).
Comparison of the InVisionSeq™ assay v1.4 and the Oncomine™ Breast assay v1): Venn diagram demonstrating the number of plasma samples with detected alternations by the two ctDNA-targeted Next-Generation Sequencing platforms InVisionSeq™ and Oncomine™ (a). Comparison of the detected alterations overall (b) and in the most affected genes ESR1 (c), PIK3CA (d) and TP53 (e) and their respective Cohen’s Kappa coefficient. Paired variant allele fractions (VAF, f) and number of detected alterations (NDA, h) according to the analysis of the 2 NGS platforms. Mann–Whitney U test p values are demonstrated. Correlation analysis for VAF (g) and NDA (i) between the 2 NGS platforms. A linear regression model has been fitted with R2 and p value demonstrated
The high congruence between the 2 platforms was also confirmed by excellent agreement across ESR1 and PIK3CA (Fig. 2b–d), with moderate agreement for TP53 (Fig. 2e). It is important to mention that due to limited plasma and resulting cfDNA for head-to-head comparison, unfortunately 7 samples had cfDNA input below the validated minimum for the InVision’s assay (and were considered indeterminate for InVision_v1.4) and therefore there is a risk of false-negative results for these samples.
In terms of VAF, of the 47 variants called, there were no statistically significant differences in the paired data (p = 0.662) or considered as a whole (p = 0.562) and high correlation was observed between the VAFs (Spearman r = 0.88 [0.99 CI 0.78, 0.93], p < 0.0001; Fig. 2f, g). The 16 variants with VAF ≥ 1% had the highest correlation (Spearman r = 0.98 [0.95 CI 0.94, 0.99], p < 0.0001), whereas the 31 variants with VAF < 1% had a lower correlation (Spearman r = 0.55 [0.95 CI 0.23, 0.71], p = 0.0013). As for the number of common detected alterations (NDAs), we found no significant differences when comparing as a whole (p = 0.235) in contrast to when comparing paired data (p = 0.013) whereas there was good correlation (Spearman r = 0.51 [0.95 CI 0.51, 0.87], p < 0.0001; Fig. 2h, i).
The alterations in genomic regions that are covered by only one assay are shown in Supplementary Table S6. Of those, 28 were represented only in the InVision panel whereas a single variant (PIK3CA p.Q546P) detected in 2 plasma samples was represented only by Oncomine (Supplementary Results).
Longitudinal analysis of plasma ctDNA in patients receiving endocrine therapy
To determine the clinical utility of mutations detected in ER-positive mBC, 8 patients with ER + mBC were included as part of a sub-study. Therapies where ctDNA sequencing was assessed were anti-estrogens, one or more aromatase inhibitors, with or without chemotherapy (exemestane, palbociclib). Two patients received fulvestrant, 2 exemestane and everolimus, 2 palbociclib and letrozole, and the remaining 2 patients received a non-steroidal aromatase inhibitor (AZD4546).
All patients whose disease progressed showed an increase in ctDNA VAF, involving at least one of PIK3CA (n = 5), GATA3 (n = 2) and/or ESR1 (n = 5). Interestingly, all PIK3CA mutations involved residues 1047 or 545 and all ESR1 mutations involved the 546–548 region, with two exceptions (p.544:L/−). Additional exon mutations were found in TP53 (3 patients), NFE2L2 (2 patients), ERBB2, KRAS, GNAS, AKT1 (one patient). Amplifications of FGFR1 or MET were detected in 2 patients. Several patients had polyclonal mutations in ESR1 (n = 2), TP53, ERBB2 or GNAS (n = 1).
Patient 1 (Pt.43; Fig. 3a) was diagnosed with ER + /PR + /HER2− invasive ductal carcinoma (IDC) with spinal metastasis. She had received Fluorouracil, epirubicin, cyclophosphamide and docetaxel (FEC-T) chemotherapy, tamoxifen, next zoladex followed by addition of fulvestrant and letrozole after worsening of liver metastases. The first research blood sample was collected while she was undergoing triple endocrine blockade, cfDNA sequencing revealed mutations in ESR1 (p.544:L/ −), GATA3 (p.317:S/FX) and PIK3CA (p.H1047R) at VAFs of 0.17%, 0.7% and 0.4%, respectively. The second research blood sample was collected 14 months later while her disease was stable. The ESR1 mutation was absent, however, the mutant VAF increased for both GATA3 (1.14%) and PIK3CA (0.86%). The patient remained stable before progressing with worsening of liver, brain and bone metastases, when GATA3 and PIK3CA mutations were detected at higher levels again and ESR1 p.544:L/- reappeared at 1.54%. As for the other markers, CA15-3 was increasing whereas CTCs were less informative and only detected at the last timepoint.
Serial monitoring of ctDNA and other blood biomarkers during endocrine therapy. Showing 2 patients with ESR1, PIK3CA, GATA3 (a) and ESR1, TP53 mutations (b) in ctDNA tracked during clinical progression. Variant allele fractions are shown as determined by sequencing. Bottom graphs show total cfDNA concentration (copies/mL), number of CTCs (per 7.5 mL of blood), CA15-3 (U/mL) and ALK-PHOS (IU/mL) for the same time points. Treatments details are given above each graph. SD stable disease, PD progressing disease, VAF variant allele frequency, CTC circulating tumour cell, CA15-3 cancer antigen 15–3, ALK-PHOS alkaline phosphatase
Patient 2 (Pt.73; Fig. 3b) was diagnosed with ER + /PR-/HER2 + IDC. She had received adjuvant tamoxifen therapy, as well as zoladex and eribulin prior to collection of the first blood sample. At this time, she began letrozole therapy but progressed with bone metastases 8 months later, when the second blood sample was collected. At that time, 2 ESR1 mutations (p.Y537C and p.D538G) were detected at VAFs of 0.16% and 1.57% respectively. The patient was enrolled into a FGFR inhibitor clinical trial for a short time before progressing once again. Despite being switched to palbociclib (with letrozole), she progressed again 2 months before the third research sample when p.Y537C and p.D538G persisted and an additional ESR1 mutation and a TP53 mutation appeared. Samples 4 and 5, were taken 1 and 2 months later at a time of further bone progression, showing ESR1 p.D538G and p.Y537S mutation at increased VAFs, an absence of p.Y537C and an emergence of a new ESR1 mutation (p.L536H) in the final sample. The patient was switched to gemcitabine and carboplatin chemotherapy, however she died 3 months after the last sample was taken. All the other markers were also increasing during disease progression.
Data for the remaining 6 patients are shown in the Supplementary Material (Supplementary Figure S1).
Discussion
This study focusses on comparison of two different NGS methods of ctDNA profiling; the InVision liquid biopsy platform, which utilizes eTAm-Seq® technology for the identification of low frequency mutations using a primer design strategy that allows for amplification of highly fragmented DNA, typical of ctDNA [9, 10]; and the Oncomine Breast Cancer panel, which is a BC-specific panel, which usesTag Sequencing technology to achieve low LOD of 0.1% for SNVs/short indels. Previous studies that used NGS platforms to compare ctDNA with tumour tissue DNA [17,18,19] demonstrated limited concordance, probably due to low tumour content in plasma and/or tumour heterogeneity.
More recent studies have compared NGS technologies for ctDNA analysis. Guardant360™ and PlasmaSELECT were compared in plasma from prostate cancer patients and showed lack of concordance in nearly 50% of patients [14]. However, these findings have been challenged, highlighting the lack of variant type or VAFs reporting and the discordance of low DNA copy numbers due to stochasticity [20, 21]. Two studies reanalysed the original data and found that indeed the alterations compared were mostly low-VAF and below the specified achievable LOD, whereas some germline variants were excluded only in one assay [22, 23]. Additionally, the cohort used was too indolent thus inappropriate for ctDNA testing [24]. In another study investigating ctDNA assay discordance, baseline plasma samples from 24 early-stage cancer patients were sent to 4 ctDNA sequencing vendors and compared with independently tested, time-matched, tumour-normal tissue pairs [15]. This orthogonal approach revealed substantial variability among the ctDNA assays with 58% discordance between matching tumour or other plasma sample, mostly observed for VAF < 1% but not > 10%, suggesting that most NGS assay discordance reflects technical variations rather than biological factors. Similarly, the Avenio ctDNA_Expanded panel and QIAseq Human_Comprehensive_Cancer panel were compared and demonstrated high coverage, sensitivity and concordance for the detection of clinically relevant variants but discordance at VAFs < 1% [25].
Our study demonstrates a high correlation between the VAFs detected, especially for variants with VAF > 1%, in agreement with Gerratana et al. who compared the NGS platforms PredicinePLUS™ and Guardant360™ in mBC [13]. As for lower VAFs, we must consider the LOD of each assay and the potential low DNA copy numbers. Although the Oncomine and the InVision assays report a LOD of 0.1% and 0.25%, respectively, Oncomine called 7 variants below the LOD (VAFs 0.05–0.09%) and InVision called a variant with VAF 0.18% in the overlapping region. In addition, 24 samples analysed only by InVision had mutations with a VAF below the LOD. Interestingly, of the 16 variants called only by Oncomine, 12 had VAFs below the LOD of the InVision assay (VAFs 0.05–0.24%). The variant TP53 p.G244C with VAF 0.46% was called by Oncomine only, however Inivata reported low copy numbers of DNA molecules for this plasma sample. As additional plasma was not available to confirm discordant variants with an orthogonal method (dPCR), it is not possible to determine whether the additional Oncomine variants represent Oncomine false-positives, InVision false-negatives or both. However, in one instance (pt.196) where Oncomine detected a TP53 mutation missed by the InVision assay (p.R282W; 0.09% VAF), InVision detected other TP53 variants at higher levels in regions not covered by Oncomine (p.251:I/X 24.1% VAF, p.V157I 4.42% VAF) indicating that the Oncomine variants could be subclonal. The Oncomine assay has called variants as low as 0.05% in another study [26] while InVision has reported detection of 88.9% of SNVs at the VAF range of 0.13–0.16% and 56.3% at 0.06–0.08% [10].
Our study fulfils all the ctDNA test concordance study design criteria [27], since the paired-sample collection was concurrent and from BC samples at progression, while the VAFs were within the detection range of both tests. Comparison between the 2 technologies demonstrated a high agreement with only 2 of 30 (6.66%) discordant samples (in both of which Inivata reported as having low input DNA), suggesting sufficient reproducibility for clinical use. When comparing the assays across the most frequently mutated genes a high agreement was observed for ESR1 and PIK3CA with low discordance rates (3.3% and 10%, respectively), comparable to other studies [13, 28] but only a moderate agreement for TP53 (20% discordance rate). Similarly, robust concordance between tissue and blood was found for PIK3CA but not TP53 mutation [29]. The majority of discordant calls occurred at VAF < 1% suggesting that detection of low-frequency mutations using NGS technologies could be limited by pre-analytical variables such as limited plasma volume, DNA isolation techniques and by sequencing artefactual errors, background noise, bioinformatics filtering thresholds and germline variant calls, resulting in false-positive discovery [15, 30]. Apart from technical reasons, biological factors such as clonal haematopoiesis of undetermined potential (CHIP)-mutations that lead to clonal expansion but not haematological neoplasia [31]- may also play a role. The detection of clonal haematopoiesis associated with KRAS, TP53, JAK2 mutations has been reported in advanced-stage NSCLC [32], so some of the low-level TP53 variants in our study might be CHIP-related.
Another part of the study was to investigate the utility of the InVision platform in mBC, since it has already proven utility in NSCLC [33,34,35]. Although the overlap of Oncomine and InVision was small (7 genes included in both assays), which is expected since InVision was designed for NSCLC, amongst those overlapping genes were genes frequently altered or implicated in the pathogenesis of mBC (PIK3CA, TP53, ESR1, ERBB2) [3]. Our results, similar to Fribbens et al. [36], demonstrate that the panel is fit-for-purpose for monitoring BC patients in a metastatic setting, since genetic alternations in many mBC-related genes were found in almost two thirds of patients. Somatic alternations were found in 20 genes and were mainly SNVs/INDELs but also few CNVs. PIK3CA, ESR1 and TP53 were the most affected genes, comparable with other studies that have used the InVision [36], Oncomine Breast Cancer [26], and PredicinePLUS™ [13] panels for hormone receptor-positive mBC.
The last part of our study was to follow mBC patients that show resistance to endocrine therapy, by analysis of longitudinal samples. There have been very few publications detailing the frequency of driver mutations in patients who have become resistant to multiple lines of endocrine therapy. Here we describe the frequent occurrence of ESR1 and PIK3CA mutations in this clinical situation. Our study may have missed other important driver genes, however, based on the panel coverage. Very low-VAF ESR1 mutations have been found in primary BC, and high-VAF in mBC, suggesting that ESR1-mutant clones are enriched by endocrine therapy [37]. A comprehensive survey of driver mutations in mBC demonstrated that the most frequent mutations associated with endocrine therapy-failure were found not only in ESR1 but also in ERBB2 and NF1 when comparing metastases with primary tumours; they found that ESR1 and ERBB2, NF1 mutations (that activate MAPKinase signalling) were mutually exclusive. The third category of mutations found in endocrine-resistant patients were the transcription factors MYC, FOXA1, TBX genes (9%), also mutually exclusive with the ESR1 (18%) and MAPKinase pathway (13%) mutations. Thus, in 60% of tumours detected mutations did not correspond to those known molecular mechanisms [38]. As regards other mutations associated with endocrine resistance in our study, KRAS [36], AKT1 [39], GATA3 [40, 41] and GNAS [21] were mutated in mBC.
One of the most interesting aspects of this study is the finding of multiple ESR1 mutations. In case new therapeutics to delay/abolish the emergence of resistance were needed, then a focus on the 536–538 codons of ESR1 and the 1047 and 545 codons of PIK3CA could be achieved by simpler/cheaper technologies including ddPCR, with some loss of sensitivity through splitting samples [36]. If inhibitors abrogating these constitutively active mutant proteins were discovered, longer remissions could be expected. However, in many patients, multiple mutations of ESR1 seem to be present, implying the need for combination therapy.
The emergence of mutations seems to coincide with an increase in other markers (CTCs, CA15-3 and ALK-PHOS) in our study, however in some cases mutations appeared before the increase of the other markers. Previously, we demonstrated that CTCs and cfDNA are more informative than the conventional biomarkers (CA15-3 and ALK-PHOS) for prediction of overall survival in mBC [42], while we and others have shown that metastatic relapse was predicted with a lead-time of up 8.9 months [2] and 6.7 months [36] by utilizing ctDNA analysis [43].
The current study has limitations (small sample size, limited material available in some cases), however it shows encouraging results on the reproducibility of 2 ctDNA-based assays performed on samples from patients in a real-world setting [44]. Our data suggest that plasma-based ctDNA sequencing seems to be reliable with both technologies tested and could be used clinically, but clinicians need to be aware of technical limitations. These platforms can be used as prognostic biomarkers for detecting molecular relapse ahead of clinical or radiologic relapse and as predictive biomarkers for predicting drug resistance and facilitate potential change of therapy.
References
Ali S, Buluwela L, Coombes RC (2011) Antiestrogens and their therapeutic applications in breast cancer and other diseases. Annu Rev Med 62:217–232. https://doi.org/10.1146/annurev-med-052209-100305
Coombes RC, Page K, Salari R, Hastings RK, Armstrong A, Ahmed S, Ali S, Cleator S, Kenny L, Stebbing J, Rutherford M, Sethi H, Boydell A, Swenerton R, Fernandez-Garcia D, Gleason KLT, Goddard K, Guttery DS, Assaf ZJ, Wu HT, Natarajan P, Moore DA, Primrose L, Dashner S, Tin AS, Balcioglu M, Srinivasan R, Shchegrova SV, Olson A, Hafez D, Billings P, Aleshin A, Rehman F, Toghill BJ, Hills A, Louie MC, Lin CJ, Zimmermann BG, Shaw JA (2019) Personalized detection of circulating tumor DNA antedates breast cancer metastatic recurrence. Clin Cancer Res 25(14):4255–4263. https://doi.org/10.1158/1078-0432.CCR-18-3663
Page K, Guttery DS, Fernandez-Garcia D, Hills A, Hastings RK, Luo J, Goddard K, Shahin V, Woodley-Barker L, Rosales BM, Coombes RC, Stebbing J, Shaw JA (2017) Next generation sequencing of circulating cell-free DNA for evaluating mutations and gene amplification in metastatic breast cancer. Clin Chem 63(2):532–541. https://doi.org/10.1373/clinchem.2016.261834
Guttery DS, Page K, Hills A, Woodley L, Marchese SD, Rghebi B, Hastings RK, Luo J, Pringle JH, Stebbing J, Coombes RC, Ali S, Shaw JA (2015) Noninvasive detection of activating estrogen receptor 1 (ESR1) mutations in estrogen receptor-positive metastatic breast cancer. Clin Chem 61(7):974–982. https://doi.org/10.1373/clinchem.2015.238717
Oxnard GR, Paweletz CP, Sholl LM (2017) Genomic analysis of plasma cell-free DNA in patients with cancer. JAMA Oncol 3(6):740–741. https://doi.org/10.1001/jamaoncol.2016.2835
Merker JD, Oxnard GR, Compton C, Diehn M, Hurley P, Lazar AJ, Lindeman N, Lockwood CM, Rai AJ, Schilsky RL, Tsimberidou AM, Vasalos P, Billman BL, Oliver TK, Bruinooge SS, Hayes DF, Turner NC (2018) Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J Clin Oncol 36(16):1631–1641. https://doi.org/10.1200/JCO.2017.76.8671
Thress KS, Brant R, Carr TH, Dearden S, Jenkins S, Brown H, Hammett T, Cantarini M, Barrett JC (2015) EGFR mutation detection in ctDNA from NSCLC patient plasma: a cross-platform comparison of leading technologies to support the clinical development of AZD9291. Lung Cancer 90(3):509–515. https://doi.org/10.1016/j.lungcan.2015.10.004
Xu T, Kang X, You X, Dai L, Tian D, Yan W, Yang Y, Xiong H, Liang Z, Zhao GQ, Lin S, Chen KN, Xu G (2017) Cross-platform comparison of four leading technologies for detecting egfr mutations in circulating tumor DNA from non-small cell lung carcinoma patient plasma. Theranostics 7(6):1437–1446. https://doi.org/10.7150/thno.16558
Gale D, Lawson ARJ, Howarth K, Madi M, Durham B, Smalley S, Calaway J, Blais S, Jones G, Clark J, Dimitrov P, Pugh M, Woodhouse S, Epstein M, Fernandez-Gonzalez A, Whale AS, Huggett JF, Foy CA, Jones GM, Raveh-Amit H, Schmitt K, Devonshire A, Green E, Forshew T, Plagnol V, Rosenfeld N (2018) Development of a highly sensitive liquid biopsy platform to detect clinically-relevant cancer mutations at low allele fractions in cell-free DNA. PLoS ONE 13(3):e0194630. https://doi.org/10.1371/journal.pone.0194630
Plagnol V, Woodhouse S, Howarth K, Lensing S, Smith M, Epstein M, Madi M, Smalley S, Leroy C, Hinton J, de Kievit F, Musgrave-Brown E, Herd C, Baker-Neblett K, Brennan W, Dimitrov P, Campbell N, Morris C, Rosenfeld N, Clark J, Gale D, Platt J, Calaway J, Jones G, Forshew T (2018) Analytical validation of a next generation sequencing liquid biopsy assay for high sensitivity broad molecular profiling. PLoS ONE 13(3):e0193802. https://doi.org/10.1371/journal.pone.0193802
Guibert N, Hu Y, Feeney N, Kuang Y, Plagnol V, Jones G, Howarth K, Beeler JF, Paweletz CP, Oxnard GR (2018) Amplicon-based next-generation sequencing of plasma cell-free DNA for detection of driver and resistance mutations in advanced non-small cell lung cancer. Ann Oncol 29(4):1049–1055. https://doi.org/10.1093/annonc/mdy005
Dhingra D, Chien R, Gu J, Brinza D, Chaudhary R, Banjara K, Li Y, Ballesteros-Villagrana E, Bramlett K (2017) Abstract 5396: An NGS workflow to detect down to 0.1% allelic frequency in cfDNA for breast and colon cancers. Cancer Res 77:5396–5396
Gerratana L, Zhang Q, Shah AN, Davis AA, Zhang Y, Wehbe F, Qiang W, Flaum L, Finkelman BS, Gradishar WJ, Platanias LC, Behdad A, Cristofanilli M (2020) Performance of a novel Next Generation Sequencing circulating tumor DNA (ctDNA) platform for the evaluation of samples from patients with metastatic breast cancer (MBC). Crit Rev Oncol Hematol 145:102856. https://doi.org/10.1016/j.critrevonc.2019.102856
Torga G, Pienta KJ (2018) Regarding the congruence between 2 circulating tumor DNA sequencing assays-reply. JAMA Oncol 4(10):1431–1432. https://doi.org/10.1001/jamaoncol.2018.2326
Stetson D, Ahmed A, Xu X, Nuttall BR, Lubinski TJ, Johnson JH, Barrett JC, Dougherty BA (2019) Orthogonal comparison of four plasma NGS tests with tumor suggests technical factors are a major source of assay discordance. JCO Precis Oncol. https://doi.org/10.1200/PO.18.00191
Page K, Guttery DS, Zahra N, Primrose L, Elshaw SR, Pringle JH, Blighe K, Marchese SD, Hills A, Woodley L, Stebbing J, Coombes RC, Shaw JA (2013) Influence of plasma processing on recovery and analysis of circulating nucleic acids. PLoS ONE 8(10):e77963. https://doi.org/10.1371/journal.pone.0077963
Chae YK, Davis AA, Jain S, Santa-Maria C, Flaum L, Beaubier N, Platanias LC, Gradishar W, Giles FJ, Cristofanilli M (2017) Concordance of genomic alterations by next-generation sequencing in tumor tissue versus circulating tumor DNA in breast cancer. Mol Cancer Ther 16(7):1412–1420. https://doi.org/10.1158/1535-7163.MCT-17-0061
Jovelet C, Ileana E, Le Deley MC, Motte N, Rosellini S, Romero A, Lefebvre C, Pedrero M, Pata-Merci N, Droin N, Deloger M, Massard C, Hollebecque A, Ferte C, Boichard A, Postel-Vinay S, Ngo-Camus M, De Baere T, Vielh P, Scoazec JY, Vassal G, Eggermont A, Andre F, Soria JC, Lacroix L (2016) Circulating cell-free tumor DNA analysis of 50 genes by next-generation sequencing in the prospective MOSCATO trial. Clin Cancer Res 22(12):2960–2968. https://doi.org/10.1158/1078-0432.CCR-15-2470
Kuderer NM, Burton KA, Blau S, Rose AL, Parker S, Lyman GH, Blau CA (2017) Comparison of 2 commercially available next-generation sequencing platforms in oncology. JAMA Oncol 3(7):996–998. https://doi.org/10.1001/jamaoncol.2016.4983
Oxnard GR, Paweletz CP (2018) Regarding the congruence between 2 circulating tumor DNA sequencing assays. JAMA Oncol 4(10):1428–1429. https://doi.org/10.1001/jamaoncol.2018.2311
Ye Q, Qi F, Bian L, Zhang SH, Wang T, Jiang ZF (2017) Circulating-free DNA mutation associated with response of targeted therapy in human epidermal growth factor receptor 2-positive metastatic breast cancer. Chin Med J (Engl) 130(5):522–529. https://doi.org/10.4103/0366-6999.200542
Agarwal N, Lanman RB, Pal SK (2018) Regarding the congruence between 2 circulating tumor DNA sequencing assays. JAMA Oncol 4(10):1429–1430. https://doi.org/10.1001/jamaoncol.2018.2317
Angiuoli SV, White JR, Jones S (2018) Regarding the congruence between 2 circulating tumor DNA sequencing assays. JAMA Oncol 4(10):1430–1431. https://doi.org/10.1001/jamaoncol.2018.2323
Rolfo C, Raez L, Cristofanilli M (2018) Regarding the congruence between 2 circulating tumor DNA sequencing assays. JAMA Oncol 4(10):1430. https://doi.org/10.1001/jamaoncol.2018.2320
Lam SN, Zhou YC, Chan YM, Foo CM, Lee PY, Mok WY, Wong WS, Fung YY, Wong KY, Huang JY, Chow CK (2020) Comparison of target enrichment platforms for circulating tumor DNA detection. Sci Rep 10(1):4124. https://doi.org/10.1038/s41598-020-60375-x
Shibayama T, Low SK, Ono M, Kobayashi T, Kobayashi K, Fukada I, Ito Y, Ueno T, Ohno S, Nakamura Y, Takahashi S (2020) Clinical significance of gene mutation in ctDNA analysis for hormone receptor-positive metastatic breast cancer. Breast Cancer Res Treat 180(2):331–341. https://doi.org/10.1007/s10549-019-05512-5
Schwaederle MC, Patel SP, Husain H, Ikeda M, Lanman RB, Banks KC, Talasaz A, Bazhenova L, Kurzrock R (2017) Utility of genomic assessment of blood-derived circulating tumor DNA (ctDNA) in patients with advanced lung adenocarcinoma. Clin Cancer Res 23(17):5101–5111. https://doi.org/10.1158/1078-0432.CCR-16-2497
O’Leary B, Hrebien S, Beaney M, Fribbens C, Garcia-Murillas I, Jiang J, Li Y, Huang Bartlett C, Andre F, Loibl S, Loi S, Cristofanilli M, Turner NC (2019) Comparison of BEAMing and droplet digital pcr for circulating tumor DNA analysis. Clin Chem 65(11):1405–1413. https://doi.org/10.1373/clinchem.2019.305805
Liang DH, Ensor JE, Liu ZB, Patel A, Patel TA, Chang JC, Rodriguez AA (2016) Cell-free DNA as a molecular tool for monitoring disease progression and response to therapy in breast cancer patients. Breast Cancer Res Treat 155(1):139–149. https://doi.org/10.1007/s10549-015-3635-5
Koessler T, Paradiso V, Piscuoglio S, Nienhold R, Ho L, Christinat Y, Terracciano LM, Cathomas G, Wicki A, McKee TA, Nouspikel T (2020) Reliability of liquid biopsy analysis: an inter-laboratory comparison of circulating tumor DNA extraction and sequencing with different platforms. Lab Investig. https://doi.org/10.1038/s41374-020-0459-7
Steensma DP, Bejar R, Jaiswal S, Lindsley RC, Sekeres MA, Hasserjian RP, Ebert BL (2015) Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood 126(1):9–16. https://doi.org/10.1182/blood-2015-03-631747
Hu Y, Ulrich BC, Supplee J, Kuang Y, Lizotte PH, Feeney NB, Guibert NM, Awad MM, Wong KK, Janne PA, Paweletz CP, Oxnard GR (2018) False-positive plasma genotyping due to clonal hematopoiesis. Clin Cancer Res 24(18):4437–4443. https://doi.org/10.1158/1078-0432.CCR-18-0143
Corradetti MN, Torok JA, Hatch AJ, Xanthopoulos EP, Lafata K, Jacobs C, Rushing C, Calaway J, Jones G, Kelsey CR, Nixon AB (2019) Dynamic changes in circulating tumor DNA during chemoradiation for locally advanced lung cancer. Adv Radiat Oncol 4(4):748–752. https://doi.org/10.1016/j.adro.2019.05.004
Guibert N, Jones G, Beeler JF, Plagnol V, Morris C, Mourlanette J, Delaunay M, Keller L, Rouquette I, Favre G, Pradines A, Mazieres J (2019) Targeted sequencing of plasma cell-free DNA to predict response to PD1 inhibitors in advanced non-small cell lung cancer. Lung Cancer 137:1–6. https://doi.org/10.1016/j.lungcan.2019.09.005
Remon J, Caramella C, Jovelet C, Lacroix L, Lawson A, Smalley S, Howarth K, Gale D, Green E, Plagnol V, Rosenfeld N, Planchard D, Bluthgen MV, Gazzah A, Pannet C, Nicotra C, Auclin E, Soria JC, Besse B (2017) Osimertinib benefit in EGFR-mutant NSCLC patients with T790M-mutation detected by circulating tumour DNA. Ann Oncol 28(4):784–790. https://doi.org/10.1093/annonc/mdx017
Fribbens C, Garcia Murillas I, Beaney M, Hrebien S, O’Leary B, Kilburn L, Howarth K, Epstein M, Green E, Rosenfeld N, Ring A, Johnston S, Turner N (2018) Tracking evolution of aromatase inhibitor resistance with circulating tumour DNA analysis in metastatic breast cancer. Ann Oncol 29(1):145–153. https://doi.org/10.1093/annonc/mdx483
Wang P, Bahreini A, Gyanchandani R, Lucas PC, Hartmaier RJ, Watters RJ, Jonnalagadda AR, Trejo Bittar HE, Berg A, Hamilton RL, Kurland BF, Weiss KR, Mathew A, Leone JP, Davidson NE, Nikiforova MN, Brufsky AM, Ambros TF, Stern AM, Puhalla SL, Lee AV, Oesterreich S (2016) Sensitive detection of mono- and polyclonal ESR1 mutations in primary tumors, metastatic lesions, and cell-free DNA of breast cancer patients. Clin Cancer Res 22(5):1130–1137. https://doi.org/10.1158/1078-0432.CCR-15-1534
Razavi P, Chang MT, Xu G, Bandlamudi C, Ross DS, Vasan N, Cai Y, Bielski CM, Donoghue MTA, Jonsson P, Penson A, Shen R, Pareja F, Kundra R, Middha S, Cheng ML, Zehir A, Kandoth C, Patel R, Huberman K, Smyth LM, Jhaveri K, Modi S, Traina TA, Dang C, Zhang W, Weigelt B, Li BT, Ladanyi M, Hyman DM, Schultz N, Robson ME, Hudis C, Brogi E, Viale A, Norton L, Dickler MN, Berger MF, Iacobuzio-Donahue CA, Chandarlapaty S, Scaltriti M, Reis-Filho JS, Solit DB, Taylor BS, Baselga J (2018) The genomic landscape of endocrine-resistant advanced breast cancers. Cancer Cell 34(3):427–438. https://doi.org/10.1016/j.ccell.2018.08.008
Rudolph M, Anzeneder T, Schulz A, Beckmann G, Byrne AT, Jeffers M, Pena C, Politz O, Kochert K, Vonk R, Reischl J (2016) AKT1 (E17K) mutation profiling in breast cancer: prevalence, concurrent oncogenic alterations, and blood-based detection. BMC Cancer 16:622. https://doi.org/10.1186/s12885-016-2626-1
Bertucci F, Ng CKY, Patsouris A, Droin N, Piscuoglio S, Carbuccia N, Soria JC, Dien AT, Adnani Y, Kamal M, Garnier S, Meurice G, Jimenez M, Dogan S, Verret B, Chaffanet M, Bachelot T, Campone M, Lefeuvre C, Bonnefoi H, Dalenc F, Jacquet A, De Filippo MR, Babbar N, Birnbaum D, Filleron T, Le Tourneau C, Andre F (2019) Genomic characterization of metastatic breast cancers. Nature 569(7757):560–564. https://doi.org/10.1038/s41586-019-1056-z
Lefebvre C, Bachelot T, Filleron T, Pedrero M, Campone M, Soria JC, Massard C, Levy C, Arnedos M, Lacroix-Triki M, Garrabey J, Boursin Y, Deloger M, Fu Y, Commo F, Scott V, Lacroix L, Dieci MV, Kamal M, Dieras V, Goncalves A, Ferrerro JM, Romieu G, Vanlemmens L, Mouret Reynier MA, Thery JC, Le Du F, Guiu S, Dalenc F, Clapisson G, Bonnefoi H, Jimenez M, Le Tourneau C, Andre F (2016) Mutational profile of metastatic breast cancers: a retrospective analysis. PLoS Med 13(12):e1002201. https://doi.org/10.1371/journal.pmed.1002201
Fernandez-Garcia D, Hills A, Page K, Hastings RK, Toghill B, Goddard KS, Ion C, Ogle O, Boydell AR, Gleason K, Rutherford M, Lim A, Guttery DS, Coombes RC, Shaw JA (2019) Plasma cell-free DNA (cfDNA) as a predictive and prognostic marker in patients with metastatic breast cancer. Breast Cancer Res 21(1):149. https://doi.org/10.1186/s13058-019-1235-8
Dawson SJ, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin SF, Dunning MJ, Gale D, Forshew T, Mahler-Araujo B, Rajan S, Humphray S, Becq J, Halsall D, Wallis M, Bentley D, Caldas C, Rosenfeld N (2013) Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med 368(13):1199–1209. https://doi.org/10.1056/NEJMoa1213261
Buono G, Gerratana L, Bulfoni M, Provinciali N, Basile D, Giuliano M, Corvaja C, Arpino G, Del Mastro L, De Placido S, De Laurentiis M, Cristofanilli M, Puglisi F (2019) Circulating tumor DNA analysis in breast cancer: Is it ready for prime-time? Cancer Treat Rev 73:73–83. https://doi.org/10.1016/j.ctrv.2019.01.004
Acknowledgements
We thank the Imperial Cancer Research UK Centre and the Imperial and Leicester Experimental Cancer Medicine Centres (ECMC), the Imperial Biomedical Research Centre (BRC) and the Imperial College Tissue Bank and the clinical teams at Charing Cross Hospital London, for supporting patient recruitment and sample collection. We would also like to thank Prof Simak Ali for his insightful discussions and critical reading of the article. We are grateful to individuals with metastatic breast cancer for participating in this study.
Funding
This study was supported by sequential programme grant funding from Cancer Research UK to J.A. Shaw and R.C. Coombes, a Clinical and Translational Research Committee Programme Award (C14315/A13462) and a Science Committee Programme Award (C14315/A23464) and by Inivata Ltd. G. Nteliopoulos is funded by the support from the Imperial NIHR Biomedical Research Centre.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
KH, WE, EG and NR are current or former employees, officers, consultants and/or share-holders of Inivata Ltd or Inivata Inc. R.C. Coombes reports speaker frees from Pfizer and has shares in Carrick Ltd. L. Kenny reports receiving speakers bureau honoraria from Pfizer, and is a consultant/advisory board member for Celgene, Novartis, and Lilly. J. Stebbing is the Editor-in-Chief of Oncogene, sat on SABs for Celltrion, Vor Biopharma and Benevolent AI, and Chairs the Board of Directors for BB Biotech Healthcare Trust and Xerion Healthcare. All the other authors declare that they have no competing interests.
Ethical Approval
This study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the Wales Multicenter Research Ethics Committee (MREC) (Imperial College Healthcare NHS Trust; Tissue Bank application number: R11015-20A; ICHTB HTA licence: 12275; REC Wales approval: 17/WA/0161).
Informed Consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Nteliopoulos, G., Page, K., Hills, A. et al. Comparison of two targeted ultra-deep sequencing technologies for analysis of plasma circulating tumour DNA in endocrine-therapy-resistant breast cancer patients. Breast Cancer Res Treat 188, 465–476 (2021). https://doi.org/10.1007/s10549-021-06220-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-021-06220-9