Abstract
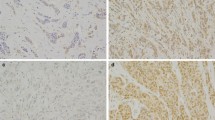
Impaired DNA damage response (DDR) may play a fundamental role in the pathogenesis of breast cancer (BC). RAD51 is a key player in DNA double-strand break repair. In this study, we aimed to assess the biological and clinical significance of RAD51 expression with relevance to different molecular classes of BC and patients’ outcome. The expression of RAD51 was assessed immunohistochemically in a well-characterised annotated series (n = 1184) of early-stage invasive BC with long-term follow-up. A subset of cases of BC from patients with known BRCA1 germline mutations was included as a control group. The results were correlated with clinicopathological and molecular parameters and patients’ outcome. RAD51 protein expression level was also assayed in a panel of cell lines using reverse phase protein array (RPPA). RAD51 was expressed in the nuclei (N) and cytoplasm (C) of malignant cells. Subcellular co-localisation phenotypes of RAD51 were significantly associated with clinicopathological features and patient outcome. Cytoplasmic expression (RAD51C+) and lack of nuclear expression (RAD51 N−) were associated with features of aggressive behaviour, including larger tumour size, high grade, lymph nodal metastasis, basal-like, and triple-negative phenotypes, together with aberrant expression of key DDR biomarkers including BRCA1. All BRCA1-mutated tumours had RAD51C+/N− phenotype. RPPA confirmed IHC results and showed differential expression of RAD51 in cell lines based on ER expression and BRCA1 status. RAD51 N+ and RAD51C+ tumours were associated with longer and shorter breast cancer-specific survival (BCSS), respectively. The RAD51 N+ was an independent predictor of longer BCSS (P < 0.0001). Lack of RAD51 nuclear expression is associated with poor prognostic parameters and shorter survival in invasive BC patients. The significant associations between RAD51 subcellular localisation and clinicopathological features, molecular subtype and patients’ outcome suggest that the trafficking of DDR proteins between the nucleus and cytoplasm might play a role in the development and progression of BC.




Similar content being viewed by others
References
Khanna KK, Jackson SP (2001) DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet 27(3):247–254
van Gent DC, Hoeijmakers JH, Kanaar R (2001) Chromosomal stability and the DNA double-stranded break connection. Nat Rev Genet 2(3):196–206
Pierce AJ, Stark JM, Araujo FD, Moynahan ME, Berwick M, Jasin M (2001) Double-strand breaks and tumorigenesis. Trends Cell Biol 11(11):S52–S59
Powell SN, Kachnic LA (2003) Roles of BRCA1 and BRCA2 in homologous recombination, DNA replication fidelity and the cellular response to ionizing radiation. Oncogene 22(37):5784–5791
Sung P (1994) Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science 265(5176):1241–1243
Baumann P, Benson FE, West SC (1996) Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell 87(4):757–766
Chen J, Silver DP, Walpita D, Cantor SB, Gazdar AF, Tomlinson G, Couch FJ, Weber BL, Ashley T, Livingston DM et al (1998) Stable interaction between the products of the BRCA1 and BRCA2 tumor suppressor genes in mitotic and meiotic cells. Mol Cell 2(3):317–328
Scully R, Chen J, Plug A, Xiao Y, Weaver D, Feunteun J, Ashley T, Livingston DM (1997) Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell 88(2):265–275
Venkitaraman AR (2001) Functions of BRCA1 and BRCA2 in the biological response to DNA damage. J Cell Sci 114(Pt 20):3591–3598
Moynahan ME, Chiu JW, Koller BH, Jasin M (1999) Brca1 controls homology-directed DNA repair. Mol Cell 4(4):511–518
Bau DT, Mau YC, Shen CY (2006) The role of BRCA1 in non-homologous end-joining. Cancer Lett 240(1):1–8
Klopfleisch R, Gruber AD (2009) Increased expression of BRCA2 and RAD51 in lymph node metastases of canine mammary adenocarcinomas. Vet Pathol 46(3):416–422
Klopfleisch R, Schutze M, Gruber AD (2010) RAD51 protein expression is increased in canine mammary carcinomas. Vet Pathol 47(1):98–101
Wiegmans AP, Al-Ejeh F, Chee N, Yap PY, Gorski JJ, Da Silva L, Bolderson E, Chenevix-Trench G, Anderson R, Simpson PT et al (2014) Rad51 supports triple negative breast cancer metastasis. Oncotarget 5(10):3261–3272
Hu J, Wang N, Wang YJ (2013) XRCC3 and RAD51 expression are associated with clinical factors in breast cancer. PLoS ONE 8(8):e72104
Sosinska-Mielcarek K, Duchnowska R, Winczura P, Badzio A, Majewska H, Lakomy J, Peksa R, Pieczynska B, Radecka B, Debska S et al (2013) Immunohistochemical prediction of brain metastases in patients with advanced breast cancer: the role of Rad51. Breast 22(6):1178–1183
Asakawa H, Koizumi H, Koike A, Takahashi M, Wu W, Iwase H, Fukuda M, Ohta T (2010) Prediction of breast cancer sensitivity to neoadjuvant chemotherapy based on status of DNA damage repair proteins. Breast Cancer Res 12(2):R17
Alshareeda AT, Negm OH, Albarakati N, Green AR, Nolan C, Sultana R, Madhusudan S, Benhasouna A, Tighe P, Ellis IO et al (2013) Clinicopathological significance of KU70/KU80, a key DNA damage repair protein in breast cancer. Breast Cancer Res Treat 139(2):301–310
Rakha EA, Putti TC, Abd El-Rehim DM, Paish C, Green AR, Powe DG, Lee AH, Robertson JF, Ellis IO (2006) Morphological and immunophenotypic analysis of breast carcinomas with basal and myoepithelial differentiation. J Pathol 208(4):495–506
Aleskandarany MA, Green AR, Benhasouna AA, Barros FF, Neal K, Reis-Filho JS, Ellis IO, Rakha EA (2012) Prognostic value of proliferation assay in the luminal, HER2-positive, and triple-negative biologic classes of breast cancer. Breast Cancer Res 14(1):R3
Rakha EA, Elsheikh SE, Aleskandarany MA, Habashi HO, Green AR, Powe DG, El-Sayed ME, Benhasouna A, Brunet JS, Akslen LA et al (2009) Triple-negative breast cancer: distinguishing between basal and nonbasal subtypes. Clin Cancer Res 15(7):2302–2310
Alshareeda AT, Rakha EA, Nolan CC, Ellis IO, Green AR (2012) Fatty acid binding protein 7 expression and its sub-cellular localization in breast cancer. Breast Cancer Res Treat 134(2):519–529
McCarty KS Jr, Miller LS, Cox EB et al (1985) Estrogen receptor analyses: correlation of biochemical and immunohistochemical methods using monoclonal antireceptor antibodies. Arch Pathol Lab Med 109:716–721
Alshareeda AT, Negm OH, Green AR, Nolan C, Tighe P, Albarakati N, Sultana R, Madhusudan S, Ellis IO, Rakha EA (2014) SUMOylation proteins in breast cancer. Breast Cancer Res Treat 144(3):519–530
Negm OH, Mannsperger HA, McDermott EM, Drewe E, Powell RJ, Todd I, Fairclough LC, Tighe PJ (2014) A pro-inflammatory signalome is constitutively activated by C33Y mutant TNF receptor 1 in TNF receptor-associated periodic syndrome (TRAPS). Eur J Immunol 44(7):2096–2110
Mannsperger HA, Gade S, Henjes F, Beissbarth T, Korf U (2010) RPPanalyzer: analysis of reverse-phase protein array data. Bioinformatics 26(17):2202–2203
Camp RL, Dolled-Filhart M, Rimm DL (2004) X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimizatio. Clin Cancer Res 10:7252–7259
Jackson SP, Bartek J (2009) The DNA-damage response in human biology and disease. Nature 461(7267):1071–1078
Benson FE, Stasiak A, West SC (1994) Purification and characterization of the human Rad51 protein, an analogue of E. coli RecA. EMBO J 13(23):5764–5771
Gupta RC, Bazemore LR, Golub EI, Radding CM (1997) Activities of human recombination protein Rad51. Proc Natl Acad Sci USA 94(2):463–468
McIlwraith MJ, Van Dyck E, Masson JY, Stasiak AZ, Stasiak A, West SC (2000) Reconstitution of the strand invasion step of double-strand break repair using human Rad51 Rad52 and RPA proteins. J Mol Biol 304(2):151–164
Graeser M, McCarthy A, Lord CJ, Savage K, Hills M, Salter J, Orr N, Parton M, Smith IE, Reis-Filho JS et al (2010) A marker of homologous recombination predicts pathologic complete response to neoadjuvant chemotherapy in primary breast cancer. Clin Cancer Res 16(24):6159–6168
Barbano R, Copetti M, Perrone G, Pazienza V, Muscarella LA, Balsamo T, Storlazzi CT, Ripoli M, Rinaldi M, Valori VM (2011) High RAD51 mRNA expression characterize estrogen receptor-positive/progesteron receptor-negative breast cancer and is associated with patient’s outcome. Int J Cancer 129(3):536–545
Barbano R, Copetti M, Perrone G, Pazienza V, Anna Muscarella L, Balsamo T, Tiziana Storlazzi C, Ripoli M, Rinaldi M, Maria Valori V (2010) High RAD51 mRNA expression characterize estrogen receptor-positive/progesteron receptor-negative breast cancer and is associated with patient’s outcome. Int J Cancer 129(3):536–545
Pedram A, Razandi M, Evinger AJ, Lee E, Levin ER (2009) Estrogen inhibits ATR signaling to cell cycle checkpoints and DNA repair. Mol Biol Cell 20(14):3374–3389
Arpino G, Weiss H, Lee AV, Schiff R, De Placido S, Osborne CK, Elledge RM (2005) Estrogen receptor–positive, progesterone receptor–negative breast cancer: association with growth factor receptor expression and tamoxifen resistance. J Natl Cancer Inst 97(17):1254–1261
Yamamori T, Meike S, Nagane M, Yasui H, Inanami O (2013) ER stress suppresses DNA double-strand break repair and sensitizes tumor cells to ionizing radiation by stimulating proteasomal degradation of Rad51. FEBS Lett 587(20):3348–3353
Mitra A, Jameson C, Barbachano Y, Sanchez L, Kote-Jarai Z, Peock S, Sodha N, Bancroft E, Fletcher A, Cooper C (2009) Overexpression of RAD51 occurs in aggressive prostatic cancer. Histopathology 55(6):696–704
Dubey A, Chouhan U (2011) Subcellular localization of proteins. Arch Appl Sci Res 3(6):392–401
Zannini L, Lecis D, Lisanti S, Benetti R, Buscemi G, Schneider C, Delia D (2003) Karyopherin-alpha2 protein interacts with Chk2 and contributes to its nuclear import. J Biol Chem 278(43):42346–42351
Huang L, Wang HY, Li JD, Wang JH, Zhou Y, Luo RZ, Yun JP, Zhang Y, Jia WH, Zheng M (2013) KPNA2 promotes cell proliferation and tumorigenicity in epithelial ovarian carcinoma through upregulation of c-Myc and downregulation of FOXO3a. Cell Death Dis 4:e745
Gorlich D, Mattaj IW (1996) Nucleocytoplasmic transport. Science 271(5255):1513–1518
Gildemeister OS, Sage JM, Knight KL (2009) Cellular redistribution of Rad51 in response to DNA damage: novel role for Rad51C. J Biol Chem 284(46):31945–31952
Stochaj U, Rassadi R, Chiu J (2000) Stress-mediated inhibition of the classical nuclear protein import pathway and nuclear accumulation of the small GTPase Gsp1p. FASEB J 14(14):2130–2132
Venkitaraman AR (2002) Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell 108(2):171–182
Shen WH, Balajee AS, Wang J, Wu H, Eng C, Pandolfi PP, Yin Y (2007) Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell 128(1):157–170
Sorensen CS, Hansen LT, Dziegielewski J, Syljuasen RG, Lundin C, Bartek J, Helleday T (2005) The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat Cell Biol 7(2):195–201
Ma CX, Janetka JW, Piwnica-Worms H (2011) Death by releasing the breaks: CHK1 inhibitors as cancer therapeutics. Trends Mol Med 17(2):88–96
Bahassi EM, Ovesen JL, Riesenberg AL, Bernstein WZ, Hasty PE, Stambrook PJ (2008) The checkpoint kinases Chk1 and Chk2 regulate the functional associations between hBRCA2 and Rad51 in response to DNA damage. Oncogene 27(28):3977–3985
Rabik CA, Dolan ME (2007) Molecular mechanisms of resistance and toxicity associated with platinating agents. Cancer Treat Rev 33(1):9–23
Burma S, Chen BP, Chen DJ (2006) Role of non-homologous end joining (NHEJ) in maintaining genomic integrity. DNA Repair (Amst) 5(9–10):1042–1048
Lieber MR (2008) The mechanism of human nonhomologous DNA end joining. J Biol Chem 283(1):1–5
Moynahan ME, Cui TY, Jasin M (2001) Homology-directed dna repair, mitomycin-c resistance, and chromosome stability is restored with correction of a Brca1 mutation. Cancer Res 61(12):4842–4850
Author information
Authors and Affiliations
Ethics declarations
Conflict of interest
The authors have no conflict of interest to declare.
Additional information
Ola H. Negm is the first joint author.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10549_2016_3915_MOESM1_ESM.docx
Supplementary material 1 (DOCX 16 kb) Sources, dilution, cut-offs point and pre-treatment conditions used of the antibodies of DNA damage sensing and repair markers used in this study
Rights and permissions
About this article
Cite this article
Alshareeda, A.T., Negm, O.H., Aleskandarany, M.A. et al. Clinical and biological significance of RAD51 expression in breast cancer: a key DNA damage response protein. Breast Cancer Res Treat 159, 41–53 (2016). https://doi.org/10.1007/s10549-016-3915-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-016-3915-8