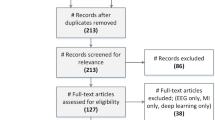
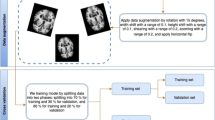
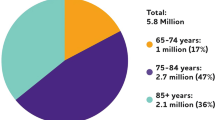Abstract
Software such as EEGLab has enabled the treatment and visualization of the tracing and cortical topography of the electroencephalography (EEG) signals. In particular, the topography of the cortical electrical activity is represented by colors, which make it possible to identify functional differences between cortical areas and to associate them with various diseases. The use of cortical topography with EEG origin in the investigation of diseases is often not used due to the representation of colors making it difficult to classify the disease. Thus, the analyses have been carried out, mainly, based on the EEG tracings. Therefore, a computer system that recognizes disease patterns through cortical topography can be a solution to the diagnostic aid. In view of this, this study compared five models of Convolutional Neural Networks (CNNs), namely: Inception v3, SqueezeNet, LeNet, VGG-16 and VGG-19, in order to know the patterns in cortical topography images obtained with EEG, in Parkinson's disease, Depression and Bipolar Disorder. SqueezeNet performed better in the 3 diseases analyzed, with Parkinson’s disease being better evaluated for Accuracy (88.89%), Precison (86.36%), Recall (91.94%) and F1 Score (89.06%), the other CNNs had less performance. In the analysis of the values of the Area under ROC Curve (AUC), SqueezeNet reached (93.90%) for Parkinson's disease, (75.70%) for Depression and (72.10%) for Bipolar Disorder. We understand that there is the possibility of classifying neurological diseases from cortical topographies with the use of CNNs and, thus, creating a computational basis for the implementation of software for screening and possible diagnostic assistance.












Similar content being viewed by others
Data Availability
Datasets generated during and/or analyzed during the current study are not publicly available due to individual privacy.
Code Availability
Not applicable.
References
Aarsland D, Creese B, Politis M, Chaudhuri KR, Ffytche DH, Weintraub D, Ballard C (2017) Cognitive decline in Parkinson disease. Nat Rev Neurol 13(4):217–231. https://doi.org/10.1038/nrneurol.2017.27
Alom MZ, Hasan M, Yakopcic C, Taha TM, Asari VK (2020) Improved inception-residual convolutional neural network for object recognition. Neural Comput Appl 32(1):279–293. https://doi.org/10.1007/s00521-018-3627-6
Alonso-Lana S, Goikolea JM, Bonnin CM, Sarró S, Segura B, Amann BL, Monté GC, Moro N, Fernandez-Corcuera P, Maristany T, Salvador R, Vieta E, Pomarol-Clotet E, McKenna PJ (2016) Structural and functional brain correlates of cognitive impairment in euthymic patients with bipolar disorder. PLoS ONE 11(7):1–17. https://doi.org/10.1371/journal.pone.0158867
Anwar AM, Eldeib AM (2020) EEG signal classification using convolutional neural networks on combined spatial and temporal dimensions for BCI systems. Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, EMBS. https://doi.org/10.1109/EMBC44109.2020.9175894
Ardakani AA, Kanafi AR, Acharya UR, Khadem N, Mohammadi A (2020) Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: results of 10 convolutional neural networks. Comput Biol Med 121(March):103795. https://doi.org/10.1016/j.compbiomed.2020.103795
Arora V, Singh M, Bhatia R (2020) Orientation-based Ant colony algorithm for synthesizing the test scenarios in UML activity diagram. Inf Softw Technol 123(February):106292. https://doi.org/10.1016/j.infsof.2020.106292
Aslan Z, Akin M (2020) Automatic detection of schizophrenia by applying deep learning over spectrogram images of EEG signals. Traitement Du Signal 37(2):235–244
Assunção WKG, Vergilio SR, Lopez-Herrejon RE (2020) Automatic extraction of product line architecture and feature models from UML class diagram variants. Inform Softw Technol. https://doi.org/10.1016/j.infsof.2019.106198
Atluri S, Frehlich M, Mei Y, Dominguez LG (2016) TMSEEG : a MATLAB-based graphical user interface for processing electrophysiological signals during transcranial magnetic stimulation. Front Neural Circuits 10(October):1–20. https://doi.org/10.3389/fncir.2016.00078
Bergstra J, Bengio Y (2012) Random search for hyper-parameter optimization. J Mach Learn Res 13:281–305
Bhandari NK, Jain M (2020) Emotion recognition and classification using Eeg: a review. Int J Sci Technol Res 9(2):1827–1836
Bhavsar R, Sun Y, Helian N, Davey N, Mayor D, Steffert T (2018) The correlation between EEG signals as measured in different positions on scalp varying with distance. Proc Comp Sci 123:92–97. https://doi.org/10.1016/j.procs.2018.01.015
Bisley JW, Goldberg ME (2010) Attention, intention, and priority in the parietal lobe. Annu Rev Neurosci 33:1–21. https://doi.org/10.1146/annurev-neuro-060909-152823
Campos F, Thorne J, Edmonds B, Schneider T, Eichele T, Debener S (2009) Clinical Neurophysiology Semi-automatic identification of independent components representing EEG artifact. Clin Neurophysiol 120(5):868–877. https://doi.org/10.1016/j.clinph.2009.01.015
Chen Z, Zhang J, Ding R, Marculescu D (2020) ViP: virtual pooling for accelerating CNN-based image classification and object detection. Proceedings - 2020 IEEE Winter Conference on Applications of Computer Vision, WACV 2020, 1169–1178. https://doi.org/10.1109/WACV45572.2020.9093418
Collazos-Huertas DF, Álvarez-Meza AM, Acosta-Medina CD, Castaño-Duque GA, Castellanos-Dominguez G (2020) CNN-based framework using spatial dropping for enhanced interpretation of neural activity in motor imagery classification. Brain Inform. https://doi.org/10.1186/s40708-020-00110-4
D’Ostilio K, Garraux G (2016) The network model of depression as a basis for new therapeutic strategies for treating major depressive disorder in Parkinson’s disease. Front Hum Neurosci 10(APR2016):1–10. https://doi.org/10.3389/fnhum.2016.00161
De Meneses FGA, Lima GD, Nunes M, Hugo Bastos V, Teixeira S (2019) Percolation theory for the recognition of patterns in topographic images of the cortical activity. Med Hypotheses. https://doi.org/10.1016/j.mehy.2019.02.021
Delorme A, Makeig S, Fabre-thorpe M, Sejnowski T (2002) From single-trial EEG to brain area dynamics. Neurocomputing 46:1057–1064
Delorme A, Makeig S (2004) EEGLAB: An open source toolbox for analysis of single-trial EEG dynamics including independent component analysis. J Neurosci Methods 134(1):9–21. https://doi.org/10.1016/j.jneumeth.2003.10.009
Dong N, Zhao L, Wu CH, Chang JF (2020) Inception v3 based cervical cell classification combined with artificially extracted features. Appl Soft Comput 93:106311. https://doi.org/10.1016/j.asoc.2020.106311
Elsaadouny M, Barowski J, Rolfes I (2020). Extracting the Features of the Shallowly Buried Objects using LeNet Convolutional Network. 14th European Conference on Antennas and Propagation, EuCAP. https://doi.org/10.23919/EuCAP48036.2020.9135701
Fatmawati E, Wijaya SK, Prawito (2018) Development prototype system of arm’s motor imagery utilizing electroencephalography signals (EEG) from emotiv with probabilistic neural network (PNN) as signal analysis. Proceedings of 2017 5th International Conference on Instrumentation, Communications, Information Technology, and Biomedical Engineering, ICICI-BME 2017, November, 179–183. https://doi.org/10.1109/ICICI-BME.2017.8537727
Foster BL, Rangarajan V, Shirer WR, Parvizi J (2015) Intrinsic and task-dependent coupling of neuronal population activity in human parietal cortex. Neuron 86(2):578–590. https://doi.org/10.1016/j.neuron.2015.03.018
Foxe JJ, Burke KM, Andrade GN, Djukic A, Frey HP, Molholm S (2016) Automatic cortical representation of auditory pitch changes in Rett syndrome. J Neurodev Disord 8(1):1–10. https://doi.org/10.1186/s11689-016-9166-5
Friedrich J, Mückschel M, Beste C (2017) Somatosensory lateral inhibition processes modulate motor response inhibition- A n EEG source localization study. Sci Rep 7(1):1–10. https://doi.org/10.1038/s41598-017-04887-z
Frolov A, Bobrov P, Biryukova E, Isaev M, Kerechanin Y, Bobrov D, Lekin A (2020) Using multiple decomposition methods and cluster analysis to find and categorize typical patterns of EEG activity in motor imagery brain-computer interface experiments. Front Robot AI. https://doi.org/10.3389/frobt.2020.00088
Funahashi S (2017) Prefrontal contribution to decision-making under free-choice conditions. Front Neurosci. https://doi.org/10.3389/fnins.2017.00431
Germann C, Marbach G, Civardi F, Fucentese SF, Fritz J, Sutter R, Pfirrmann CWA, Fritz B (2020) Deep convolutional neural network-based diagnosis of anterior cruciate ligament tears: performance comparison of homogenous versus heterogeneous knee MRI cohorts with different pulse sequence protocols and 15-T and 3-T magnetic field strengths. Investigat Radiol 55(8):499–506. https://doi.org/10.1097/RLI.0000000000000664
Ghassemi N, Shoeibi A, Rouhani M (2020) Deep neural network with generative adversarial networks pre-training for brain tumor classification based on MR images. Biomed Signal Process Control 57:101678. https://doi.org/10.1016/j.bspc.2019.101678
Gore E, Rathi S (2019) Surveying machine learning algorithms on eeg signals data for mental health assessment. 2019 IEEE Pune Section International Conference, PuneCon 2019, 1–6. https://doi.org/10.1109/PuneCon46936.2019.9105749
Gorji A, Speckmann EJ (2009) Epileptiform EEG spikes and their functional significance. Clin EEG Neurosci 40(4):230–233. https://doi.org/10.1177/155005940904000404
Grosselin F, Navarro-Sune X, Raux M, Similowski T, Chavez M (2018) CARE-rCortex: a Matlab toolbox for the analysis of CArdio-REspiratory-related activity in the Cortex. J Neurosci Methods 308(August):309–316. https://doi.org/10.1016/j.jneumeth.2018.08.011
Grouiller F, Thornton RC, Groening K, Spinelli L, Duncan JS, Schaller K, Siniatchkin M, Lemieux L, Seeck M, Michel CM, Vulliemoz S (2011) With or without spikes: localization of focal epileptic activity by simultaneous electroencephalography and functional magnetic resonance imaging. Brain 134(10):2867–2886. https://doi.org/10.1093/brain/awr156
Guo X, Zhang HBRL, Ding X, Tian R (2018) Attention-Based Combination of CNN and RNN (Vol. 1). Springer International Publishing. https://doi.org/10.1007/978-3-030-04212-7
Hasnain M, Pasha MF, Ghani I, Imran M, Alzahrani MY, Budiarto R (2020) Evaluating trust prediction and confusion matrix measures for web services ranking. IEEE Access 8:90847–90861. https://doi.org/10.1109/ACCESS.2020.2994222
Hassanpour M, Malek H (2020) Learning document image features with SqueezeNet convolutional neural network. Int J Eng Trans A 33(7):1201–1207. https://doi.org/10.5829/ije.2020.33.07a.05
Helm K, Viol K, Weiger TM, Tass PA, Grefkes C, Del Monte D, Schiepek G (2018) Neuronal connectivity in major depressive disorder: a systematic review. Neuropsychiatr Dis Treat 14:2715–2737. https://doi.org/10.2147/NDT.S170989
Hooi LS, Nisar H, Voon YV (2016) Comparison of motion field of EEG topo-maps for tracking brain activation. IECBES 2016 - IEEE-EMBS Conference on Biomedical Engineering and Sciences, pp 251–256. https://doi.org/10.1109/IECBES.2016.7843452
Iandola FN, Han S, Moskewicz MW, Ashraf K, Dally WJ, Keutzer K (2016) SqueezeNet: AlexNet-level accuracy with 50x fewer parameters and <0.5MB model size. 1–13. http://arxiv.org/abs/1602.07360
Imtiaz SA, Rodriguez-Villegas E (2015) An open-source toolbox for standardized use of PhysioNet Sleep EDF expanded database. Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, EMBS, 2015-November, 6014–6017. https://doi.org/10.1109/EMBC.2015.7319762
Jadhav P, Rajguru G, Datta D, Mukhopadhyay S (2020) Automatic sleep stage classification using time–frequency images of CWT and transfer learning using convolution neural network. Biocybernet Biomed Eng 40(1):494–504. https://doi.org/10.1016/j.bbe.2020.01.010
Jung TP, Humphries C, Lee TW, McKeown MJ, Iragui V, Makeig, & Sejnowski, T. J. (2000a) Removing electroencephalographic artifacts from by blind source separation. Psychophysiology 37(2):163–178
Jung T, Makeig S, Westerfield M, Townsend J, Courchesne E, Sejnowskp TJ (2000a) Analyzing and visualizing single-trial event-related potentials
Kim HC, Kang MJ (2020) Comparison of hyper-parameter optimization methods for deep neural networks. J IKEEE 24(4):969–974. https://doi.org/10.7471/ikeee.2020.24.4.969
Khaleghi A, Sheikhani A, Mohammadi MR, Nasrabadi AM (2015) Evaluation of cerebral cortex function in clients with bipolar mood disorder I (BMD I) compared with BMD II using QEEG analysis. Iran J Psychiatry 10(2):93–99
Khan AU, Akram M, Daniyal M, Zainab R (2019) Awareness and current knowledge of Parkinson’s disease: a neurodegenerative disorder. Int J Neurosci 129(1):55–93. https://doi.org/10.1080/00207454.2018.1486837
Klonovs J, Petersen C, Olesen H, Hammershoj A (2013) ID proof on the go: development of a mobile EEG-based biometric authentication system. IEEE Veh Technol Mag 8(1):81–89. https://doi.org/10.1109/MVT.2012.2234056
Koenig T, Prichep L, Lehmann D, Sosa PV, Braeker E, Kleinlogel H, Isenhart R, John ER (2002) Millisecond by millisecond year by year : normative EEG microstates and developmental stages. Neuroimage 48:41–48. https://doi.org/10.1006/nimg.2002.1070
Kwon SK, Jung HS, Baek WK, Kim D (2017) Classification of forest vertical structure in south korea from aerial orthophoto and lidar data using an artificial neural network. Appl Sci 7(10). https://www.mdpi.com/2076-3417/7/10/1046
LeCun Y, Bottou L, Bengio Y, Haffner P et al (1998) Gradient-based learning applied to document recognition. Proc IEEE 86(11):2278–2324. https://doi.org/10.1109/5.726791
Li L, Ma L, Jiao L, Liu F, Sun Q, Zhao J (2020a) Complex contourlet-CNN for polarimetric SAR image classification. Pattern Recogn 100:107110. https://doi.org/10.1016/j.patcog.2019.107110
Li T, Jin D, Du C, Cao X, Chen H, Yan J, Chen N, Chen Z, Feng Z, Liu S (2020b) The image-based analysis and classification of urine sediments using a LeNet-5 neural network. Compt Methods Biomech Biomed Eng 8(1):109–114. https://doi.org/10.1080/21681163.2019.1608307
Ma F, Sun T, Liu L, Jing H (2020) Detection and diagnosis of chronic kidney disease using deep learning-based heterogeneous modified artificial neural network. Futur Gener Comput Syst 111:17–26. https://doi.org/10.1016/j.future.2020.04.036
Maletic V, Raison C (2014) Integrated neurobiology of bipolar disorder. Front Psych. https://doi.org/10.3389/fpsyt.2014.00098
Makeig S, Jung TP, Bell AJ, Ghahremani D, Sejnowski TJ (1997) Blind separation of auditory event-related brain responses into independent components. Proc Natl Acad Sci USA 94(20):10979–10984. https://doi.org/10.1073/pnas.94.20.10979
Mak LE, Minuzzi L, MacQueen G, Hall G, Kennedy SH, Milev R (2017) The default mode network in healthy individuals: a systematic review and meta-analysis. Brain Connect 7(1):25–33. https://doi.org/10.1089/brain.2016.0438
Makeig S, Westerfield M, Jung T, Covington J, Townsend J, Sejnowski TJ, Courchesne E (1999) Functionally independent components of the late positive event-related potential during visual spatial attention. J Neurosci 19(7):2665–2680
Martinez P, Lopez J, Rodriguez FJ, Wiggins JB, Boyer KE (2020) Novice debugging in block-based and hybrid environments. Ann Conf Innovat Technol Comput Sci Educat ITiCSE 1(1):1291. https://doi.org/10.1145/1122445.1122456
Michalopoulos K, Bourbakis N (2013). Microstate analysis of the EEG using local global graphs. 13th IEEE International Conference on BioInformatics and BioEngineering, IEEE BIBE 2013, https://doi.org/10.1109/BIBE.2013.6701583
Michalopoulos K, Bourbakis N (2014a) Using dynamic bayesian networks for modeling EEG topographic sequences. 2014 36th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, EMBC 2014, 4928–4931. https://doi.org/10.1109/EMBC.2014.6944729
Mishra A et al (2019) Noise Removal in EEG Signals Using SWT–ICA Combinational Approach. (2019). Department of Electronics and Communication Engineering, Shri Ramswaroop Memorial Group of Professional Colleges (SRMGPC), Lucknow 226028, Uttar Pradesh, India, pp 217–224
Mzurikwao D, Williams Samuel O, Grace Asogbon M, Li X, Li G, Yeo WH, Efstratiou C, Siang Ang C (2019) A channel selection approach based on convolutional neural network for multi-channel EEG motor imagery decoding. Proceedings - IEEE 2nd International Conference on Artificial Intelligence and Knowledge Engineering, AIKE 2019, pp 195–202. https://doi.org/10.1109/AIKE.2019.00042
Narin A, Kaya C, Pamuk Z (2020) Department of biomedical engineering, zonguldak bulent ecevit university, 67100, Zonguldak, Turkey. https://arxiv.org/abs/2003.10849
Pal A, Gautam AK, Singh YN (2015) Evaluation of bioelectric signals for human recognition. Proc Compt Sci 48:746–752. https://doi.org/10.1016/j.procs.2015.04.211
Palaniappan R, Mandic DP (2007) EEG based biometric framework for automatic identity verification. J VLSI Signal Process Syst Signal Image Video Technol 49(2):243–250. https://doi.org/10.1007/s11265-007-0078-1
Peña CJ, Bagot RC, Labonté B, Nestler EJ (2014) Epigenetic signaling in psychiatric disorders. J Mol Biol 426(20):3389–3412. https://doi.org/10.1016/j.jmb.2014.03.016
Perrotta A, Pais-Vieira C, Allahdad MK, Bicho E, Pais-Vieira M (2020) Differential width discrimination task for active and passive tactile discrimination in humans. MethodsX 7:8. https://doi.org/10.1016/j.mex.2020.100852
Planas E, Cabot J (2020) How are UML class diagrams built in practice? A usability study of two UML tools: magicdraw and papyrus. Compt Standards Interfaces 67:103363. https://doi.org/10.1016/j.csi.2019.103363
Priyanshu A et al (2021) Efficient hyperparameter optimization for differentially private deep learning pp. 1–5. http://arxiv.org/abs/2108.03888
Raghu S, Sriraam N, Temel Y, Rao SV, Kubben PL (2020) EEG based multi-class seizure type classification using convolutional neural network and transfer learning. Neural Netw 124:202–212. https://doi.org/10.1016/j.neunet.2020.01.017
Rattay F (1993) Simulation of artificial neural reactions produced with electric fields. Simul Pract Theory 1(3):137–152. https://doi.org/10.1016/0928-4869(93)90003-9
Sairamya NJ, George ST, Ponraj DN, Subathra MSP (2018) Detection of epileptic EEG signal using improved local pattern transformation methods. Circuits Syst Signal Process 37(12):5554–5575. https://doi.org/10.1007/s00034-018-0829-1
Scaini G, Valvassori SS, Diaz AP, Lima CN, Benevenuto D, Fries GR, Quevedo J (2020) Neurobiology of bipolar disorders: a review of genetic components, signaling pathways, biochemical changes, and neuroimaging findings. Braz J Psychiatry 42(5):536–551. https://doi.org/10.1590/1516-4446-2019-0732
Shang R, He J, Wang J, Xu K, Jiao L, Stolkin R (2020) Dense connection and depthwise separable convolution based CNN for polarimetric SAR image classification. Knowl-Based Syst 194:105542. https://doi.org/10.1016/j.knosys.2020.105542
Shah SAA, Zhang L, Bais A (2020) Dynamical system based compact deep hybrid network for classification of Parkinson disease related EEG signals. Neural Netw 130:75–84. https://doi.org/10.1016/j.neunet.2020.06.018
Shalbaf A, Bagherzadeh S, Maghsoudi A (2020) Transfer learning with deep convolutional neural network for automated detection of schizophrenia from EEG signals. Phys Eng Sci Med. https://doi.org/10.1007/s13246-020-00925-9
She Q, Hu B, Gan H, Fan Y, Nguyen T, Potter T, Zhang Y (2018) Safe semi-supervised extreme learning machine for EEG signal classification. IEEE Access 6:49399–49407. https://doi.org/10.1109/ACCESS.2018.2868713
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. https://arxiv.org/abs/1409.1556v6
Solé-Casals J, Caiafa CF, Zhao Q, Cichocki A (2018) Brain-computer interface with corrupted EEG data: a tensor completion approach. Cogn Comput 10(6):1062–1074. https://doi.org/10.1007/s12559-018-9574-9
Srivastava S, Kumar P, Chaudhry V, Singh A (2020) Detection of ovarian cyst in ultrasound images using fine-tuned VGG-16 deep learning network. SN Compt Sci 1(2):1–8. https://doi.org/10.1007/s42979-020-0109-6
Stropahl M, Bauer AKR, Debener S, Bleichner MG (2018) Source-modeling auditory processes of EEG data using EEGLAB and brainstorm. Front Neurosci 12(MAY):1–11. https://doi.org/10.3389/fnins.2018.00309
Sun Y, Xue B, Zhang M, Yen GG, Lv J (2020) Automatically designing CNN architectures using the genetic algorithm for image classification. IEEE Trans Cybernet 50(9):3840–3854. https://doi.org/10.1109/TCYB.2020.2983860
Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z (2016) Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 2818–2826. https://doi.org/10.1109/CVPR.2016.308
Taqi AM, Al-azzo F, Mariofanna M, Al-Saadi JM (2017) Classification and discrimination of focal and non-focal EEG signals based on deep neural network. International Conference on Current Research in Computer Science and Information Technology (ICCIT), Slemani Iraq, pp 86–92. https://doi.org/10.1109/CRCSIT.2017.7965539
Ucar F, Korkmaz D (2020) COVIDiagnosis-Net: Deep Bayes-SqueezeNet based diagnosis of the coronavirus disease 2019 (COVID-19) from X-ray images. Med Hypotheses 140(April):109761. https://doi.org/10.1016/j.mehy.2020.109761
Usakli AB (2010) Improvement of EEG signal acquisition: an electrical aspect for state of the Art of front end. Comput Intell Neurosci. https://doi.org/10.1155/2010/630649
Wang J (2020) A deep learning approach for atrial fibrillation signals classification based on convolutional and modified Elman neural network. Futur Gener Comput Syst 102:670–679. https://doi.org/10.1016/j.future.2019.09.012
Wang Z, Chen C, Li W, Yuan W, Han T, Sun C, Tao L, Zhao Y, Chen W (2018) A multichannel EEG acquisition system with novel Ag NWs/PDMS flexible dry electrodes. Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, EMBS, pp 1299–1302. https://doi.org/10.1109/EMBC.2018.8512563
Wendling F, Bartolomei F, Senhadji L (2009) Spatial analysis of intracerebral electroencephalographic signals in the time and frequency domain: identification of epileptogenic networks in partial epilepsy. Philosoph Trans Royal Soc 367(1887):297–316. https://doi.org/10.1098/rsta.2008.0220
Whitfield-Gabrieli S, Ford JM (2012) Default mode network activity and connectivity in psychopathology. Annu Rev Clin Psychol. https://doi.org/10.1146/annurev-clinpsy-032511-143049
Xiao J, Wang J, Cao S, Li B (2020) Application of a novel and improved VGG-19 network in the detection of workers wearing masks. J Phys. https://doi.org/10.1088/1742-6596/1518/1/012041
Xu J, Zhang Y, Miao D (2020a) Three-way confusion matrix for classification: a measure driven view. Inf Sci 507:772–794. https://doi.org/10.1016/j.ins.2019.06.064
Xu M, Yao J, Zhang Z, Li R, Yang B, Li C, Li J, Zhang J (2020b) Learning EEG topographical representation for classification via convolutional neural network. Pattern Recogn. https://doi.org/10.1016/j.patcog.2020.107390
Yang ES et al (2017) Hyperparameter tuning for hidden unit conditional random fields. Eng Comput 34(6):2054–2062. https://doi.org/10.1108/EC-11-2015-0350
Zhang P, Wang X, Chen J, You W, Zhang W (2019a) Spectral and temporal feature learning with two-stream neural networks for mental workload assessment. IEEE Trans Neural Syst Rehabil Eng 27(6):1149–1159. https://doi.org/10.1109/TNSRE.2019.2913400
Zhang P, Wang X, Zhang W, Chen J (2019b) Learning spatial-spectral-temporal EEG features with recurrent 3D convolutional neural networks for cross-task mental workload assessment. IEEE Trans Neural Syst Rehabil Eng 27(1):31–42. https://doi.org/10.1109/TNSRE.2018.2884641
Acknowledgements
The authors would like to thank the Brazilian National Council for Scientific and Technological Development (CNPq) and State of Maranhão Research Funding Agency (FAPEMA).
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors report no conflict of interest.
Additional information
Handling Editor: Dezhong Yao .
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
de Meneses, F.G.A., Teles, A.S., Nunes, M. et al. Neural Networks to Recognize Patterns in Topographic Images of Cortical Electrical Activity of Patients with Neurological Diseases. Brain Topogr 35, 464–480 (2022). https://doi.org/10.1007/s10548-022-00901-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10548-022-00901-4