Abstract
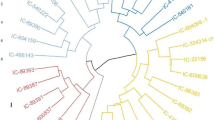
The present study describes the assessment of genetic diversity and relationships among 79 Job’s tears (Coix lacrymajobi L.) accessions collected from China and Korea using 17 microsatellite markers. A total of 57 alleles were detected with an average of 3.4 alleles per locus. A high frequency of rare alleles (36.3 %) was observed within the collection. Values for observed (HO), expected heterozygosity (HE) and Shannon’s information index (I) within the analysis ranged from 0.00 (GBssrJT183) to 0.81 (GBssrJT130), from 0.01 (GBssrJT170) to 0.65 (GBssrJT130) and from 0.034 (GBssrJt170) to 1.13 (GBssrJT130), respectively. The locus GBJT130 was the most informative marker with the highest values for observed and effective alleles as well as for HO, HE and I. Based on the UPGMA algorithm, the majority of the Chinese accessions grouped in one cluster, whereas all the Korean accessions grouped together in a separate cluster, indicating that Chinese accessions are genetically quite distinct from Korean accessions. No relation between genetic relatedness among Job’s tears accessions and their place of collection was observed. Chinese accessions exhibited greater within population polymorphism (P = 95 %, HE = 0.30, I = 0.52) than the accessions from Korea (P = 68 %, HE = 0.13, I =0.24), indicating their potentiality as a reservoir of novel alleles for crop improvement. However, in general the low diversity within each population indicates a narrow genetic base within our collection.
Similar content being viewed by others
Abbreviations
- AFLP:
-
amplified fragment length polymorphism
- PCR:
-
polymerase chain reaction
- RAPD:
-
random amplification of polymorphic DNA
- RFLP:
-
restriction fragment length polymorphism
- SSRs:
-
simple sequence repeats
References
Arber, A.: The Gramineae. A Study of Cereal, Bamboo, and Grass. — Wheldon & Wesley, New York 1965.
Boys, J., Cherry, M., Dayanandan, S.: Microsatellite analysis reveals genetically distinct populations of red pine (Pinus resinosa, Pinaceae). — Amer. J. Bot. 92: 833–841, 2005.
Budak, H., Pedraza, F., Cregan, P.B., Baenziger, P.S., Dweikat, I.: Development and utilization of SSRs to estimate the degree of genetic relationships in a collection of Pearl millet germplasm. — Crop Sci. 43: 2284–2290, 2003.
Coburn, J.R., Temnykh, S.V., Paul, E.M., McCouch, S.R.: Design and application of microsatellite marker panels for semi automated genotyping of rice (Oryza sativa L.). — Crop Sci. 42: 2092–2099, 2002.
Depeiges, A., Goubely, C., Lenoir, A., Cocherel, S., Picard, G., Raynal, M., Grellet, F., Delseny, M.: Identification of the most represented repeated motifs in Arabidopsis thaliana microsatellite loci. — Theor. appl. Genet. 91: 160–168, 1995.
Dice, L.R.: Measures of the amount of ecologic association between species. — Ecology 26: 297–302, 1945.
Dikshit, H.K., Jhang, T., Singh, N.K. Koundal, K.R., Bansal, K.C., Chandra, N., Tickoo, J.L., Sharma, T.R.: Genetic differentiation of Vigna species by RAPD, URP and SSR markers. — Biol. Plant. 51: 451–457, 2007.
Dje, Y., Heuertz, M., Lefebvre, C., Vekemans, X.: Assessment of genetic diversity within and among germplasm accessions in cultivated sorghum using microsatellite markers. — Theor. appl. Genet. 100: 918–925, 2000.
Gao, L.-Z.: Microsatellite variation within and among populations of Oryza officinalis (Poaceae), an endangered wild rice from China. — Mol. Ecol. 14: 4287–4297, 2005.
Li, X., Huang, Y., Li, J., Corke, H.: Characterization of genetic variation and relationships among Coix germplasm accessions using RAPD markers. — Genet. Resour. Crop Evolut. 48: 189–194, 2001.
Lopes, M.S., Sefc, K.M., Eiras Dias, E., Steinkellner, H., Camara Machado, L.M., Camara Machado, A.: The use of microsatellites for germplasm management in a Portuguese grapevine collection. — Theor. appl. Genet. 99: 733–739, 1999.
Ma, K.H., Kim, K.H., Dixit, A., Yu, J.W., Chung, J.W., Lee, J.H., Cho, E.G., Kim, T.S., Park, Y.J.: Newly developed polymorphic microsatellite markers in Job’s tears (Coix lacryma-jobi L.). — Mol. Ecol. Notes 6: 689–691, 2006.
Matus, I.A., Hayes, P.M.: Genetic diversity in three groups of barley germplasm assessed by simple sequence repeats. — Genome 45: 1095–1106, 2002.
Mohan, M., Nair, S., Krishna, T.G., Yano, M., Rhodes, I.: Genome mapping, molecular markers and marker-assisted selection in crop plants. — Mol. Breed. 3: 87–103, 1997.
Narasimhan, S., Padmesh, P., Nair, G.M.: Assessment of genetic diversity in Coscinium fenestratum. — Biol. Plant. 50: 111–113, 2006.
Orti, G., Pearse, D.E., Avise, J.C.: Phylogenetic assessment of length variation at a microsatellite locus. — Proc. nat. Acad. Sci. USA 94: 10745–10749, 1997.
Pritchard, J.K., Stephens, M., Donnelly, P.: Inference of population genetic structure using multilocus genotype data. — Genetics 155: 945–959, 2000.
Pritchard, J.K., Wen, W.: Documentation for Structure Software, Ver. 2. — Department of Human Genetics, the University of Chicago, Chicago 2003.
Qin, F., Li, J., Li, X., Corke, H.: AFLP and RFLP linkage map in Coix. — Genet. Resour. Crop Evolut. 52: 209–214, 2005.
Rohlf, F.J.: NTSYS-pc: Numerical Taxonomy and Multivariate Analysis System, Version 2.0. — Exeter Publishing, Setauket — New York 1998.
Roussel, V., Koenig, J., Beckert, M., Balfourier, F.: Molecular diversity in French bread wheat accessions related to temporal trends and breeding programs. — Theor. appl. Genet. 108: 920–930, 2004.
Schuelke, M.: An economic method for the fluorescent labeling of PCR fragments. — Natur. Biotechnol. 18: 233–234, 2000.
Yeh, F.C., Yang, R.C., Boyle, T.J.B.: Popgene. Version 1.31. Microsoft Window-based Freeware for Population Genetic Analysis. — University of Alberta and Centre for International Forestry Research, Edmonton 1999.
Yifru, T., Hammer, K., Huang, X.Q., Roder, M.S.: Regional patterns of microsatellite diversity in Ethiopian tetraploid wheat accessions. — Plant Breed. 125: 125–130, 2006.
Zhou, H.F., Xie, Z.W., Ge, S.: Microsatellite analysis of genetic diversity and population genetic structure of a wild rice (Oryza rufipogon Griff.) in China. — Theor. appl. Genet. 107: 332–339, 2003.
Acknowledgements
This study was supported by the BioGreen 21 project (#20080401034058) of the Rural Development Administration (RDA), Republic of Korea.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ma, K.H., Kim, K.H., Dixit, A. et al. Assessment of genetic diversity and relationships among Coix lacryma-jobi accessions using microsatellite markers. Biol Plant 54, 272–278 (2010). https://doi.org/10.1007/s10535-010-0047-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10535-010-0047-6