Abstract
Ecological research has focused on the importance of environmental factors on spatial biodiversity variations and organisation. This is important because of scant conservation resources. We used stepwise backward selection and random feature selection (RFE) to identify a parsimonious model that can predict species richness and diversity metrics in response to three models; biotic, abiotic, and topo-edaphic. Our results show that both metrics are good predictors of one another, mainly because species diversity is a combination of species richness and abundance, and further highlights the importance of biotic variables in predicting species distribution. The two modelling techniques selected soil texture and its interactions with topographic variables as the most important variables. However, random forest performed worse than multiple linear regression in the prediction of diversity metrics. This research highlights the importance of topographically controlled edaphic factors as drivers of species richness and diversity in mountainous grasslands where topography inherently controls the geomorphic, hydrological, and, as a result, ecological processes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Many environmental factors affect plant species richness and diversity in grassland communities (Auestad et al. 2008). Habitat fragmentation and loss are causing significant declines in grassland biodiversity, and ecosystem services are dwindling (Zulka et al. 2014). Ecological, topographic, and management intensity are the most important features influencing grassland biodiversity (Orlandi et al. 2016). However, in the Afromontane grasslands, these features are many and site-specific. Thus it is difficult for conservation management. Conservation resources are often scarce, and knowledge about which environmental drivers promote species diversity and richness in mountainous protected areas could allow for the prioritisation of the hotspots and allocation of conservation efforts (Olea et al. 2010). For example, nutrient-poor grasslands in Central Europe are identified as one of the biodiversity hotspots in the region and, therefore, highly valued by conservationists (Becker and Brändel 2007).
Vegetation response to environmental factors should be studied at specific yet multiple scales(Reitalu et al., 2012). Variations in grassland vegetation result from complex gradients of soil moisture and element concentrations locally and climatic variables regionally (Auestad et al. 2008). Research on vegetation-environment relationships is scanty for most parts of the Afromontane grasslands, especially locally; hence, conservation efforts may be haphazard and ad-hoc. For example, some edaphic variables, such as soil carbon content, increased species richness, while heavy metals and the C/N ratio were unrelated and decreased species richness, respectively (Becker and Brändel 2007). Grassland landscapes can be better understood when good predictors of species richness are used (Zulka et al. 2014). Predictors can be used to comprehend the complex patterns of species richness, especially in mountainous grasslands vulnerable to global environmental changes.
In alpine grassland, environmental factors, such as climate, topography and soil, are pivotal for maintaining plant diversity—for example, altitude and slope influence species composition. Inherently, topography in mountainous areas determines temperature, elevation, and hydrology; therefore, it is an important determinant of vascular plant diversity (Moeslund et al. 2013). In addition, topography in mountainous habitats facilitates geomorphological processes such as erosion and, thus, soil fertility along slope gradients, as well as aspects affecting ecological processes. Ultimately, this interplay of environmental factors influences vegetation variations in mountainous areas and can be used to identify biodiversity hotspots for direct conservation protocols in protected areas (Lee and Chun 2016). Mountain ranges in Southern Africa vary in topography and aspect, providing suitable habitats for various plants and animals (Brown and du Preez 2020). The role of topography needs to be investigated, especially in habitats where topographic interplays are poorly understood (Moeslund et al. 2013).
The most active research in ecology has been to understand the importance of environmental factors on spatial biodiversity variations and organisation. Species distribution models, which use a combination of species occurrence or abundance and ecological aspects, can aid in gaining insight into vegetation distribution across landscapes and allow for extrapolation in space and time. Furthermore, key modelling steps such as gathering data, selecting relevant predictors, and appropriate modelling algorithms can influence the robustness and realism of the model (Elith and Leathwick 2009). Modelling algorithms such as parametric generalised linear models and non-parametric random forest (RF) can be used to assess the uncertainty of modelling algorithms (Bittner et al. 2011). In protected areas of developing countries, such as Golden Gate Highlands National Park in South Africa, conservation resources are limited and should be used sparingly. Therefore, good models that can predict the potential distribution of vascular plant diversity linked to nutrient-rich forage can give initial insights into animal distribution and forage preference. Subsequently, precise carrying capacity models can be developed. This research aimed to gain insights into the main drivers of vascular plant diversity in the mountainous grassland of Golden Gate Highlands National Park by two different modelling techniques.
Methods
Study area
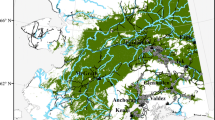
The study was conducted at Golden Gate Highlands National Park, in the North-Eastern part of the Free State province, South Africa (Fig. 1). The park comprises 32,758.35 ha and lies between 28°27’ S – 28°37’ S and 28°33’ E – 28°42’ E. The Park is located in mountainous grasslands at the foothills of the Drakensberg and forms part of the mesic highveld grassland with marked variation in geology, topography, and rainfall. The following soil types are identified in the park: shallow rocky soils (Glenrosa and Mispah), deep soil along drainage lines (Oakleaf), well-developed sand soils (Hutton and Clovelly), and clayey structured soils (Milkwood and Tambakulu) (SANParks 2020). The park is characterised by summer rainfall, temperate summers, and cold winters. The rainfall season stretches from September to April, with a mean annual rainfall ranging from 800 mm to 2,000 mm (Kay et al. 1993). The park lies between 1,892 m and 2,829 m above sea level and, hence, comprises the following grasslands units: Eastern Free State Sandy grasslands (Gm 4), Basotho Montane Shrubland (Gm 5), Lesotho Highveld Basalt Grassland (Gd 8), and Northern Drakensberg Highveld (Gd5) (Mucina and Rutherford 2006).
Data collection
Field data collection
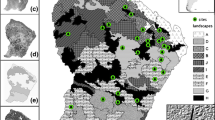
The park was stratified into relatively homogenous physiographic-physiognomic units (Kay et al. 1993). Vegetation sampling was undertaken in a 30 × 30 m plot size randomly placed within homogeneous patches of grass communities (Fig. 2). Four 30 m transects were placed horizontally at every 10 m interval within the plot, the plot where generated using the “generate random points” tool in ArcMap 10.7. In each plot, a 1 × 1 m quadrat size was placed at every 10 m interval along the transect for vascular plant species identification and ariel visual cover estimations. In total, 142 plots with 16 quadrats were sampled in the study area. Data were collected in March, April, and May 2019, which are the rainy growing months for South African montane grasslands; this area receives late rains for summer, thus prolonged vegetation growing season, in addition to the xeromorphic characteristics of plant species. In each quadrat, all vascular plant species of the standing vegetation were identified to species level where possible. The 1 × 1 m quadrat was gridded into 100 cells of 1 cm2 each, representing 1% cover to record an aerial cover of each species Fig. 3.
Environmental variables and fire severity
Various environmental predictor variables from different data sets were used to measure and model their influence on species richness and diversity. For soil, both chemical and physical variables were used; bulk density (BD), silt fragments (SF), pH, coarse fragments (CF), soil organic carbon (SOC), sand (SD), and nitrogen of topsoil (15 cm) were downloaded from International Soil Reference and Information Center (https://soilgrids.org). The park comprises shallow rocky soils, and while field sampling is encouraged in underrepresented areas (Hengl et al. 2017), physical soil sampling may be damaging in this park. Using this grided soil data with relatively good accuracies (Hengl et al. 2017) is non-destructive, therefore, ideal for a conservation area. The elevation data, i.e. SRTM DEM at 30 m resolution, was obtained from US Geological Survey’s EROS data centre (https://earthexplorer.usgs.gov). The slope was derived from DEM using the Spatial Analyst Tool in ArcGIS 10. 4. Topographic and edaphic variables were multiplied to determine the topo-edaphic variables. Fire severity data were acquired from a study conducted in the area (Adagbasa et al. 2018). The study estimated fire severity using the Normalized Burn Ratio (NBR) index by analysing a pre/post-fire season (April – September 2017) from remote sensing images with burnt and unburnt pixels.
Diversity metrics
Species diversity was calculated using the Shannon diversity index H’=-pi*lnpi where pi is the proportion (species cover) of each species in the quadrat; this analysis was computed using the vegan package (Oksanen et al. 2012) in R studio (R core Team, 2022) Furthermore, total species richness was measured by counting all vascular plants species that were recorded for each quadrat. Subsequently, all the quadrat values were averaged to attain the plot level value of each diversity metric.
Data analysis
Simple multiple linear regression
Simple multiple linear regression was used to examine the relationship between diversity metrics (species diversity and richness) and a set of three models of explanatory environmental variables. The biotic model included diversity metrics as predictor variables amongst abiotic-based variables. The abiotic model had bulk density, silt fragments, sand, pH, soil organic content, coarse fragments, elevation, slope, fire severity, and soil nitrogen. The topo-edaphic model included interactions between topographic and soil variables. The diversity metrics (species diversity and richness) were used as both response and predictor variables because of the high colinearity between the two. Subsequently, a backward selection was used to eliminate redundant variables and select the optimal variables explaining species richness and diversity from the set of environmental variables based on the lowest Akaike’s Information Criterion. Statistical analyses were performed using R statistical software (R Core Team, 2022).
Random forest
A set of biotic, abiotic, and topo-edaphic variables was used to input the nonparametric RF method to predict species richness and diversity. Random forest is a highly recommended method for ecologists and remote sensing scientists and was developed to improve classification and regression trees (CART) by using a large set of regression trees (Ramoelo et al. 2015). The ‘random forest’ package in R statistical software was used to analyse the data. Optimising the number of variables required to predict species richness and diversity was determined using a random feature selection (RFE) based on leave-one-out cross-validation, and the root mean square error (RMSE). The RFE is a simple backward selection algorithm implemented via the ‘caret’ package in R statistical software (R Core Team, 2022).
Model performance
The statistical measure of model precision and robustness, the r-squared (R2) and root mean square error (RMSE), was determined to test the performance of the modelling algorithms using the selected stepwise and RFE variables in predicting the diversity metrics. The parameters were used to assess the strength of the relationship between the observed and predicted species richness and diversity.
Results
The species and richness diversity metrics had a coefficient of variation of 25% and 39%, respectively (Table 1). All edaphic variables had a very low coefficient of variation (< 10%), except for coarse fragments (38%) and soil organic content (32%). For topographic variables, the slope had the highest (80%) and elevation with the lowest (0.06%) coefficient of variation. Fire severity had a coefficient variability of 26%. All the topo-edaphic variables exhibited a substantially high coefficient of variation (> 82%).
The optimal environmental variables explaining species richness and diversity in grassland communities in Golden Gate Highland National Park are given in Table 2. The step-wise backward selection retained species diversity, silt fragments, and fire severity as the optimal variables explaining species richness. However, species diversity had a significantly positive (7.04, p < 0.05), while silt fragments and fire severity had a significantly (p < 0.05) negative relationship (-0.24 and − 0.41, respectively). Bulk density (-0.02) and silt fragments (-0.03) were identified as the two optimal abiotic variables with a significantly (p < 0.05) negative relationship with species richness. Similarly, in the topo-edaphic model, the stepwise selected elevation, soil bulk density, silt fragments, sand, and pH had a significantly negative (p < 0.05) albeit minute relationship with species richness. Species diversity was optimally explained by species richness (0.10), soil pH (0.02), and fire severity (0.04). Coarse fragments had a significantly (p < 0.05) negative relationship (-0.03) with species diversity amongst the selected abiotic variables. However, two topographic variables were identified as optimally explaining species diversity, namely elevation interaction with soil bulk density and sand, but with very small estimate coefficients.
Measures of model precision and robustness (R2 and RMSE) to test the performance of the modelling algorithms in predicting species richness and diversity in grassland communities of Golden Gate Highland National Park are given in Table 3. The top 5 variables selected within the biotic model by RFE were species diversity (RMSE = 2.22, R2 = 0.60), silt fragments (RMSE = 2.06, R2 = 0.65), elevation (RMSE = 2.12, R2 = 0.65), and (RMSE = 2.14, R2 = 0.65) and coarse fragments (RMSE = 2.06, R2 = 0.67). Within the abiotic model, elevation (RMSE = 3.27, R2 = 0.12), and (RMSE = 3.36, R2 = 0.09), silt fragments (RMSE = 3.26, R2 = 0.13), soil organic content (RMSE = 3.26, R2 = 0.14), and bulk density (RMSE = 3.79, R2 = 0.02) were the top 5 variables explaining species richness. An interaction between elevation and bulk density (RMSE = 0.56, R2 = 0.97) was identified as the top variable within the topo-edaphic model to explain species richness. Species richness (RMSE = 0.24, R2 = 0.64) and elevation (RMSE = 0.26, R2 = 0.59) were selected as the top two variables explaining species diversity within the biotic model. However, within the abiotic-based model, sand (RMSE = 0.45, R2 = 0.09), elevation (RMSE = 0.38, R2 = 0.12), soil organic content (RMSE = 0.38, R2 = 0.09), silt fragments (RMSE = 0.41, R2 = 0.03), and bulk density (RMSE = 0.45, R2 = 0.37) were the top five most important variables. Lastly, elevation-bulk density (RMSE = 0.28, R2 = 0.54) and slope-bulk density (RMSE = 0.26, R2 = 0.59) interactions were the two most important variables within the topo-edaphic model.
The multiple linear regression models (Fig. 4) performed better than the RF models (Fig. 5). The multiple linear regression biotic models explained 72% of the variation in species richness with an RMSE of 1.84, while abiotic and topo-edaphic models explained 12% (RMSE = 3.06) and 99% (RMSE = 0.11), respectively. However, the models’ performances were reduced when species diversity was the response variable. The biotic model explained 71% variation (RMSE = 0.12). On the other hand, abiotic and topo-edaphic models explained 0.8% and 69% variation, respectively. The RF biotic model explained 62% of the variation in species richness, with the abiotic model explaining 0.03 (RMSE = 3.46) and the topo-edaphic model 91% (RMSE = 1.20). When species diversity was the response variable, the biotic model explained 59% of the variation, the abiotic model − 0.2% (RMSE = 0.45) and topo-edaphic 64% (RMSE = 0.26).
Discussion
Soil edaphic variables exhibited a very low coefficient of variation (< 10%), while slope showed a very high variation of 80%, second to all the topo-edaphic variables with a variation of > 82%. The low variation of the soil edaphic dataset in Golden Gate Highland National Park can be attributed to unvarying land-use activities, both current (protected area with grazing activities) and historic (farming: crop cultivation) because soil stoichiometry differs amongst and is influenced by land-use activities. For example, soil total carbon (C) and nutrients varied among different grassland types and land use in Alpine Qinghai-Tibetan Plateau (Han et al. 2019). Furthermore, a mean slope of 6.4% that ranged from completely flat (0.0%) to gentle (0.26%) was observed, despite the high variation in the dataset, which may have been because of the difficulty of steep sampling slopes > 45 degrees.
The diversity metrics served as both response and predictor variables. This gives insight into forces controlling species richness and diversity in response to biotic and abiotic factors (Tilman 1993). The high collinearity between the two metrics was inevitable because species richness comprises the number of species, and the Shannon-Wiener diversity index considers both species richness and abundance. Despite having similar traits, the diversity metrics may differ in their sensitivity to biotic and abiotic factors. For example, species richness may be sensitive to rapid changes in vital rare species, while species diversity may react to the changes of abundances dominant species. Thus, Symstad and Jonas (2011) suggested that “the response of richness and diversity to drivers reflect different changes in the plant communities”.
This research showed a significantly negative relationship between species richness, silt fragments, and fire severity. pH and fire severity, however, influenced species diversity via the stepwise algorithm in the biotic model. Edaphic factors are related to geomorphic heterogeneity, mainly slope stability, which can affect species richness at specific scales in mountainous areas (Malanson et al. 2020). Moreover, fire severity is one of the variables selected by the RF to explain species richness negatively. High fire severity will cause more physical/structural damage to the grassland vegetation community; hence vegetation seldom fully recovers to pre-fire conditions (Adagbasa et al. 2020). Generally, however, variations in fire regimes can serve as a stimulant for seedling establishment and emergence (Olea et al. 2010), while high fire severity, in particular, can obliterate existing species in grassland plant ecosystems. Interestingly, elevation and edaphic interaction variables were identified as optimal for influencing species richness and diversity. This emphasises the role of topography in influencing local plant diversity. For instance, elevation may limit spatial seed distribution, especially by animals that find it difficult to ascend steep slopes (Moeslund et al. 2013). The RF algorithm similarly identified silt fragments, elevation, and other soil texture variables important for explaining species richness. This also shows that topographically controlled edaphic factors such as silt fragments can be drivers of species richness in mountainous areas (Filibeck et al. 2019). Therefore, it is essential to prioritise soil conservation in protected areas to improve plant species diversity, especially in humid mountainous regions susceptible to erosion and nutrient leaching.
Species richness and diversity were affected differently by biotic, abiotic, and topo-edaphic models. As all the multiple linear regression algorithms better predicted such, species richness compared to species diversity. The RF performed worse in predicting diversity metrics because of the high prediction error, i.e. RMSE. Our results concur with studies that question the predictive power of RF in estimating vegetation parameters(Filibeck et al. 2019; Kosicki 2020), mainly because it is a non-parametric technique that does not make assumptions about statistical distribution (Ramoelo et al. 2015). However, RF is recommended for ecological use because many variables can be tested, are robust to multicollinearity, and are independent of any assumption; in comparison, stepwise multiple regression is prone to multicollinearity and overfitting, while machine learning methods are prone to different specific conditions in an ecosystem and lack analytical assumptions, which influence their predictive power.
Conclusion
Conservation of natural resources warrants active research to gain insight into the distribution of species occurrence, abundance, and composition. Because conservation resources are dwindling with heightened threats to biodiversity, our study can provide a framework for identifying environmental drivers of species richness through the selection of parsimonious models. The two metrics were useful rangeland indicators. When species distribution is well understood, they can be used to model carrying capacity because they are linked to plant productivity and nutrition. Furthermore, the importance of topographically controlled edaphic grasslands as drivers of species richness and diversity in mountainous grasslands is highlighted. Topography inherently controls the geomorphic and ecological processes. Fire severity is one of the most concerning predictors of species diversity to park managers, which is susceptible to sporadic anthropogenic fire; drivers of fire severity ought to be investigated for abating species loss due to high fire severity. The findings show that diversifying model techniques can help disentangle species richness’s complex patterns.
References
Adagbasa GE, Adelabu SA, Okello TW (2018) ‘Spatio-temporal assessment of fire severity in a protected and mountainous ecosystem’, International Geoscience and Remote Sensing Symposium (IGARSS), 2018-July(December), pp. 6572–6575. https://doi.org/10.1109/IGARSS.2018.8518268
Adagbasa EG, Adelabu SA, Okello TW (2020) ‘Development of post-fire vegetation response-ability model in grassland mountainous ecosystem using GIS and remote sensing’, ISPRS Journal of Photogrammetry and Remote Sensing, 164(September 2019), pp. 173–183. https://doi.org/10.1016/j.isprsjprs.2020.04.006
Auestad I, Rydgren K, Økland RH (2008) Scale-dependence of vegetation‐environment relationships in semi‐natural grasslands. J Veg Sci 19(1):139–148. https://doi.org/10.3170/2007-8-18344
Becker T, Brändel M (2007) Vegetation-environment relationships in a heavy metal-dry grassland complex. Folia Geobotanica 42(1):11–28. https://doi.org/10.1007/BF02835100
Bittner T et al (2011) Comparing modelling approaches at two levels of biological organisation - climate change impacts on selected Natura 2000 habitats. J Veg Sci 22(4):699–710. https://doi.org/10.1111/j.1654-1103.2011.01266.x
Brown LR, du Preez J (2020) Alpine Vegetation of Temperate Mountains of Southern Africa. Encyclopedia of the World’s biomes. Elsevier, pp 395–404. https://doi.org/10.1016/B978-0-12-409548-9.11892-5.
Elith J, Leathwick JR (2009) Species distribution models: ecological explanation and prediction across space and time. Annu Rev Ecol Evol Syst 40(1):677–697. https://doi.org/10.1146/annurev.ecolsys.110308.120159
Filibeck G et al (2019) Exploring the drivers of vascular plant richness at very fine spatial scale in sub-mediterranean limestone grasslands (Central Apennines, Italy). Biodivers Conserv 28(10):2701–2725. https://doi.org/10.1007/s10531-019-01788-7
Han Y et al (2019) ‘Response of soil nutrients and stoichiometry to elevated nitrogen deposition in an alpine grassland on the Qinghai-Tibetan Plateau’, Geoderma, 343(September 2018), pp. 263–268. doi:https://doi.org/10.1016/j.geoderma.2018.12.050
Hengl T et al (2017) ‘SoilGrids250m: Global gridded soil information based on machine learning’, PLOS ONE. Edited by B. Bond-Lamberty, 12(2), p. e0169748. https://doi.org/10.1371/journal.pone.0169748
Kay C, Bredenkamp GJ, Theron GK (1993) The plant communities of the Golden Gate Highlands National Park in the north-eastern Orange Free State. South Afr J Bot 59(4):442–449. https://doi.org/10.1016/S0254-6299(16)30717-7
Kosicki JZ (2020) Generalised Additive Models and Random Forest Approach as effective methods for predictive species density and functional species richness. Environ Ecol Stat 27(2):273–292. https://doi.org/10.1007/s10651-020-00445-5
Lee C-B, Chun J-H (2016) Environmental drivers of patterns of Plant Diversity along a wide environmental gradient in korean temperate forests. Forests 7(12):19. https://doi.org/10.3390/f7010019
Malanson GP et al (2020) Alpine plant community diversity in species–area relations at fine scale. Arct Antarct Alp Res 52(1):41–46. https://doi.org/10.1080/15230430.2019.1698894
Moeslund JE et al (2013) Topography as a driver of local terrestrial vascular plant diversity patterns. Nord J Bot 31(2):129–144. https://doi.org/10.1111/j.1756-1051.2013.00082.x
Mucina L, Rutherford MC (2006) ‘The vegetation of South Africa, Lesotho and Swaziland.’, Strelitzia 19, (December), pp. 1–30. Available at: http://ebooks.cambridge.org/ref/id/CBO9781107415324A009
Oksanen AJ et al (2012) Community Ecology Package, Comprehensive R Archive Network. Available at: http://mirror.bjtu.edu.cn/cran/web/packages/vegan/vegan.pdf
Olea PP, Mateo-Tomás P, de Frutos Á (2010) ‘Estimating and Modelling Bias of the Hierarchical Partitioning Public-Domain Software: Implications in Environmental Management and Conservation’, PLoS ONE. Edited by S. Plaistow, 5(7), p. e11698. https://doi.org/10.1371/journal.pone.0011698
Orlandi S et al (2016) Environmental and land use determinants of grassland patch diversity in the western and eastern Alps under agro-pastoral abandonment. Biodivers Conserv 25(2):275–293. https://doi.org/10.1007/s10531-016-1046-5
Ramoelo A et al (2015) Monitoring grass nutrients and biomass as indicators of rangeland quality and quantity using random forest modelling and WorldView-2 data. Int J Appl Earth Obs Geoinf 43:43–54. https://doi.org/10.1016/j.jag.2014.12.010
Reitalu, T. et al. (2012) ‘Responses of grassland species richness to local and landscape factors depend on spatial scale and habitat specialization’, Journal of Vegetation Science. Edited by J. Fridley, 23(1), pp. 41–51. https://doi.org/10.1111/j.1654-1103.2011.01334.x.
R Core Team (2022). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/.
SANParks (2020) Golden Gate Highlands National Park Park Management Plan. South African National Parks
Symstad AJ, Jonas JL (2011) Incorporating Biodiversity into Rangeland Health: Plant Species Richness and Diversity in Great Plains Grasslands. Rangel Ecol Manage 64(6):555–572. https://doi.org/10.2111/REM-D-10-00136.1
Tilman D (1993) Species richness of experimental Productivity gradients: how important is colonization limitation? Ecology 74(8):2179–2191. https://doi.org/10.2307/1939572
Zulka KP et al (2014) Species richness in dry grassland patches of eastern Austria: a multi-taxon study on the role of local, landscape and habitat quality variables. Agric Ecosyst Environ 182:25–36. https://doi.org/10.1016/j.agee.2013.11.016
Funding
Open access funding provided by University of the Free State.
Author information
Authors and Affiliations
Contributions
All author’s conceived and designed the research; Katlego K Mashiane performed the fieldwork, data mining, and analysis; Katlego K Mashiane wrote and edited the manuscript; Abel Ramoelo and Samuel Adelabu reviewed the manuscripts.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that no financial interests/personal relationships may be considered potential competing interests. Data used in this manuscript is available and can be provided upon request.
Additional information
Communicated By Daniel Sanchez Mata
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mashiane, K.K., Ramoelo, A. & Adelabu, S. Diversifying modelling techniques to disentangle the complex patterns of species richness and diversity in the protected afromontane grasslands. Biodivers Conserv 32, 1423–1436 (2023). https://doi.org/10.1007/s10531-023-02560-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10531-023-02560-8