Abstract
Characterizing genetic diversity and structure of populations is essential for the effective conservation of threatened species. Orchis patens sensu lato is a narrowly distributed tetraploid species with a disjunct distribution (i.e., Northern Italy, North Africa and the Canary Islands), which is facing a severe decline. In this study, we evaluated levels of genetic diversity and population structuring using 12 new nuclear microsatellite markers. Our analyses of genetic differentiation based on multiple approaches (Structure analysis, PCA analysis, and F-statistics using the ploidy-independent Rho-index) showed that gene flow is low across the range of O. patens s.l., particularly in the Canary Islands. Clear differences in allele frequencies between Italy, Algeria and the Canary Islands underlie the genetic differentiation retrieved. Our study provides support for the recognition of O. canariensis as a sister species to O. patens and the separation of the Italian populations as a new subspecies of O. patens. Despite the high heterozygosity values found in all populations (ranging from 0.4 to 0.7), compatible with the tetraploid status of the species, small population sizes and reduced gene flow will be likely detrimental for the different populations in the long term, and we recommend immediate conservation actions to counteract further fragmentation and population decline.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
As we are facing the sixth age of extinction (Swarts and Dixon 2009; Barnosky et al. 2011), an increasing number of species are threatened by decreased population sizes, habitat degradation and habitat fragmentation. Endangered species are in most cases characterised by small, fragmented and declining populations (Brooks et al. 2002). Such limited groups of individuals are at high risk of extinction owing to decreased genetic diversity and the growing effect of biological stochasticity (Fahrig 2003; Henle et al. 2004). There is strong evidence, indeed, that genetic factors, such as low genetic diversity and consequently reduced reproductive fitness, are driving threatened taxa to extinction (Spielman et al. 2004; Vellend and Geber 2005; Garner et al. 2020). Preserving genetic diversity is therefore one of the essential requirements to ensure population survival (Hoban et al. 2020; Laikre et al. 2020), especially under anthropogenic and environmental pressures, including climate change (Hewitt 2004; Jump et al. 2009). Rare species with low genetic diversity are more vulnerable to environmental and biological stresses compared to species with higher genetic diversity (Leimu et al. 2006; Honnay and Jacquemyn 2007; Aguilar et al. 2008), and small, isolated and declining populations of rare species, under climate change, may also become maladapted (Borrell et al. 2020). Habitat fragmentation or the complete loss of the habitat are major reasons for depletion of genetic variation in plants (Young et al. 1996; Frankham et al. 2002). Generally, species with narrow distribution ranges or small and/or isolated populations are more affected by loss of their natural habitat, and they are more susceptible to extinction due to depletion of genetic diversity than those species that are widely distributed and have relatively large, non-fragmented populations (Hamrick and Godt 1996; Frankham et al. 2002; Pandey et al. 2015), although life-history traits may play an important role in buffering genetic diversity loss (Honnay and Jacquemyn 2007).
Spatial distribution of genetic variation within a species may offer useful insights into fundamental ecological and environmental processes and information important for identifying distinct genetic groups (if present) across its range (Allendorf and Luikart 2007; Mkare et al. 2017; Gargiulo et al. 2019a). The delineation of conservation units for an endangered species is critical not only for their long-term survival, but also for directing the prioritization of conservation efforts (Petit et al. 1998; Schwartz et al. 2007; Volkmann et al. 2014; Mkare et al. 2017; Médail and Baumel 2018). Characterizing genetic diversity and structure of populations is useful in informing assisted gene flow (Borrell et al. 2020), or to identify high priorities for effective conservation of threatened species (Fay 2018; Väli et al. 2019). This is even more urgent for orchids, as approximately half of the 1641 currently assessed in the IUCN Red List are in the categories Vulnerable, Endangered and Critically Endangered (Fay 2020; IUCN, 2020).
The genus Orchis, which includes about 20–25 species, is divided into two subgenera, Orchis and Masculae, with the first commonly called the “anthropomorphic group” (Kretzschmar et al. 2007). The topology of Bateman et al. (2003) shows the non-anthropomorphic Orchis species are well-supported as monophyletic. Cytological examinations of different Orchis taxa have revealed a main chromosome number of 2n = 42 (e.g. Mrkvicka 1992; Aedo and Herrero 2005), but exceptions with aberrant values have been found in section Robustocalcare (2n = 40; Hautzinger 1978), which includes the only tetraploid species in the genus, O. patens. However, more recent counts indicated tetraploid O. patens with 2n = 84 (Pridgeon et al. 2001; Bernardos et al. 2006).
Orchis patens sensu lato is a species with a disjunct geographic range, with O. patens subsp. patens occurring in Italy (Liguria region) and North Africa (Algeria and Tunisia), and O. patens subsp. canariensis occurring in the Canary Islands (Orsenigo et al. 2016). Several authors have considered O. canariensis (syn. O. patens subsp. canariensis) and O. patens (syn. O. patens subsp. patens) as sister species (e.g., Bateman et al. 2003; Bernardos et al. 2006), unlike WCSP (2020), in which O. canariensis is considered as the basionym of O. patens subsp. canariensis. Although not resolving relationships in O. patens s.l., ITS-based phylogenetic analyses showed that O. patens s.l. is the sister group of O. spitzelii (Bateman et al. 2003), a species predominantly found in Mediterranean regions, including northern Africa and western Asia, with outlying populations on the island of Gotland. A recent study on seed micromorphology of O. patens subsp. patens, however, identified some similarities with the O. mascula group (Calevo et al. 2017), a group of taxa distributed in Europe, north-western Africa and western Asia, thus opening up the hypothesis of an allotetraploid origin. Orchis patens is assessed in the Italian (Orsenigo et al. 2016) and European IUCN Red Lists (Rankou 2011) as Endangered (EN), and in the Mediterranean IUCN Red List (Calevo et al. 2018) as Vulnerable (VU), but all the assessments agree that, given the clear morphological differences between subspecies and the lack of studies investigating genetic diversity, clarifying taxonomic uncertainties at species and subspecies levels should be a priority for the conservation of O. patens s.l.
Moreover, although O. patens is reported as the only tetraploid species of the genus (Mrkvicka 1992; Pridgeon et al. 2001), it is not clear whether different cytotypes (ploidy levels) occur (Bernardos et al. 2006) and codominant markers, especially microsatellites, may provide indirect information through the allele counts observed (Besnard and Baali-Cherif 2009; Gompert and Mock 2017). In this study, we designed new nuclear microsatellite markers to elucidate genetic structure and diversity in O. patens s.l., which will aid in the development of appropriate conservation strategies. In particular, our aims were to i) ascertain the occurrence of gene flow among populations, with a focus on indirectly detecting differences in ploidy; ii) evaluate the infraspecific delimitation and taxonomic rank of O. patens subsp. patens and O. patens subsp. canariensis; iii) verify the potential use and informativity of the markers herein developed in other congeneric species.
We refer to Canarian populations as O. canariensis and to African and Italian populations as O. patens s.s. from here on.
Materials and methods
Plant material
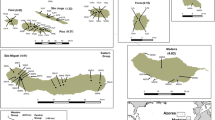
We collected leaf samples from populations of both subspecies, representative of the distribution range of the species. Samples of O. patens s.s. were collected in north-western Italy (in figures referred to as OP samples, Fig. 1a and d), in four localities of the Liguria region (16 samples from Romaggi, 11 samples from Breccanecca, 12 samples from Portofino and six samples from Capreno), and in two localities of Algeria (Fig. 1b and e): ten samples from Ait Zikki, in the Bejaia region (in figures referred to as OPAZ samples), and ten samples from Ain Talhi, in the Souk-Ahras region (in figures referred to as OPSA samples), carefully avoiding natural hybrids. Samples of O. canariensis (Fig. 1c and f) were collected from Tenerife (ten samples, in figures referred to as TN), Gran Canaria (ten samples, in figures to referred as GC) and La Gomera (nine samples, in figures referred to as LG). Collection of plant material was performed after requesting permission from the competent authorities and respecting regional rules.
Inflorescences and leaves of Orchis patens s.s. from Italy (a, d) and Algeria (b, e), and O. canariensis from Gran Canaria (c) and La Gomera (f). It is possible to notice differences in morphology and colours. In particular, leaves in O. canariensis (f) are larger and more rounded than those lanceolate of the subspecies patens (d, e). Furthermore, leaves in the Italian populations (d) present pale spots that are instead absent in the African populations (e), the one described by Desfontain (1799). Pictures by Jacopo Calevo (a, c, d), Khellaf Rebbas (b, e) and Ángel B. Fernandez (f)
DNA extraction and Illumina sequencing
Total genomic DNA was extracted from silica-dried leaf tissues using the DNeasy Plant Mini Kit (QIAGEN) according to the manufacturer's instructions, after disruption with TissueLyser. The quantity and quality of DNA samples were assessed by spectrophotometry (ND-1000 Spectrophotometer NanoDropH; Thermo Scientific, Wilmington, Germany) and by electrophoresis on a 1% agarose gel.
A genome-skimming approach (Straub et al. 2012; Dodsworth 2015) was used for the detection of microsatellites (Viruel et al. 2018; Landoni et al. 2020). Genomic libraries were prepared using NEBNext® UltraTM II DNA Library Prep Kit for Illumina® (New England Biolabs, Ipswich, MA, United States) with AMPure XP magnetic beads for size selection (300–350 bp) and NEBNext® Multiplex Oligos for Illumina® (Dual Index Primer Sets I and II) as barcodes for simultaneous sequencing (Viruel et al. 2019). Library quality was evaluated using a QuantusTM fluorometer (Promega Corp.) and an Agilent 4200 TapeStation (Agilent Technologies, Santa Clara, CA, United States). Multiplexed libraries were then sequenced on an Illumina MiSeq (Illumina, Inc.) lane. Trimmomatic v0.35 (Bolger et al. 2014) was used to remove low-quality reads and adapter sequences flagged by FastQC v0.11.7 (Andrews 2010).
Microsatellite design and analysis
Microsatellites were identified and primers designed with MSATCOMMANDER 1.0.8 (Faircloth 2008); duplicate reads and those containing more than one SSR array were manually checked and removed. Primers fulfilling the following criteria were selected: optimal size 18–22 bp, not directly flanking the SSR motif, lacking ambiguous bases, low self- and pair-product complementary parameters, amplicon expected size < 300 bp, and melting temperature (Tm) difference < 1.5 °C (Viruel et al. 2018).
Preliminary screening was carried out for 40 nuclear primer pairs on a 2% agarose gel. After testing and optimization of primers with a subset of samples, the final set of markers consisted of 12 microsatellite primer pairs (Table 1);other primer pairs were discarded due to issues with reproducibility and PCR product yield, possibly resulting from the tetraploid status of the species. Representative sequences for the microsatellite sequences were deposited in GenBank (accession numbers MT799099-MT799110).
PCR mix was prepared by adding 6 μL of (2 ×) DreamTaq PCR MasterMix (ThermoFisher Scientific), 0.5 μL of 0.4% (w/v) bovine serum albumin (BSA), 0.5 μL of each primer (10 μM), 1 μL of template DNA (ca. 10 ng/μL), and H2O up to a final volume of 10 μL. Amplifications were performed with the following PCR conditions: 4 min of initial denaturation at 94 °C followed by 25 to 35 cycles (see Table 1 for primer details) of 30 secs of denaturation at 94 °C, 30 secs of annealing (see Table 1 for temperature details), 45 secs of elongation at 72 °C and an extra extension of 10 min at 72 °C, with the exception for primer pairs Opat_27 for which it was necessary to perform a touch-down PCR by decreasing annealing temperature of 0.2 °C at each cycle (see Table 1). One μL of PCR product was added to 10 μL HiDi™ formamide (Applied Biosystems) and 0.15 μL 500 ROX Size Standard (Applied Biosystems) and capillary electrophoresis was run on an ABI3730 DNA Analyzer (Applied Biosystems).
Fragment sizing was performed in GeneMapper v.5 (Applied Biosystems) and peaks were visually checked before assigning final allele sizes. We inferred the minimum chromosome number from the maximum number of peaks detected; this procedure has been shown to give results consistent to flow cytometry (Besnard and Baali-Cherif 2009; Stark et al. 2011; Nemorin et al. 2013; Teixeira et al. 2014; Donkpegan et al. 2015; Gompert and Mock 2017). We used the R package polysat v1.7 (Clark and Jasieniuk 2011; Clark and Schreier 2017) to explore segregation patterns in our microsatellite loci. Understanding whether loci are polysomic or are inherited as separate subgenomes can give some clues about the origin of polyploidy in O. patens and, most importantly, can inform about the correct treatment of the dataset for subsequent analyses (Dufresne et al. 2014; Hardy 2016; Meirmans et al. 2018; Meirmans and Liu 2018). To avoid the bias induced by population structure, we kept individuals from Italy, Algeria and the Canary Islands as three separate populations before performing the analyses in polysat; we used the function processDatasetAllo with default parameters and excluding the locus Opat_38 (see results).
To check the transferability of the primers hereby developed, we also tested them in five other ‘non-anthropomorphic’ Orchis taxa, O. provincialis, O. mascula subsp. mascula, O. mascula subsp. ichnusae, O. spitzelii, and O. × fallax (O. patens × O. provincialis), and four anthropomorphic species, O. anthropophora, O. simia, O. militaris and O. purpurea.
Genetic diversity and structure
As we did not have prior information about the extent of gene flow in O. patens s.l., we conducted the analyses both by grouping individuals based on geographical regions (i.e., Italy, Algeria and the Canary Islands) and on sampling localities (i.e., nine sampling localities across the three regions).
Standard population genetic parameters such as mean number of alleles (Num), effective number of alleles (Eff_num), observed heterozygosity (HO), gene diversity (HS) and inbreeding coefficient GIS based on the 12 nuclear microsatellites were estimated using the program GenoDive v.3 (Meirmans and Van Tienderen 2004; Meirmans 2020). In addition, we performed a hierarchical analysis of molecular variance (AMOVA; Meirmans 2012) based on the Rho-statistics (ρ-statistics), which is ploidy-independent (Ronfort et al. 1998). All analyses in GenoDive were performed by applying the correction for unknown dosage of the alleles. Test for deviations from Hardy–Weinberg proportions was not attempted since it was not possible to record allelic dosage information for all the loci (Meirmans et al. 2018; Gargiulo et al. 2019b). We used the programme SPAGeDI v.1.5 (Hardy and Vekemans 2002) to produce a matrix of pairwise Rho values testing significance with 20,000 permutations; in addition, we estimated the selfing rate per locality using the standardized identity disequilibrium (Hardy 2016). Monomorphic loci were excluded from the computations.
We further analyzed genetic structure using different approaches. In polysat, we constructed a neighbor-joining tree based on a square dissimilarity matrix and we performed a principal component analysis (PCA) based on Bruvo distances (Bruvo et al. 2004). The model-based clustering procedure implemented in Structure v2.3.4 (Pritchard et al., 2000) was used to evaluate the number of different genetic clusters, K. We applied the Recessive Alleles model (Falush et al. 2007) to handle missing allelic dosage information. Preliminary analyses were conducted both with the Admixture model and the Non-Admixture model, testing 1 < K < 8. The final analysis was conducted with K ranging from 1 to 8, with a 5 × 105 burn-in period, 5 × 105 MCMC runs, and ten iterations per each K value, using the Non-Admixture model for the total dataset, and the Admixture model for subsets based on sampling locations and infraspecific delimitations. The most likely value of K was evaluated using ΔK (Evanno et al. 2005) and the estimators introduced by Puechmaille (2016) for uneven datasets (MedMedK, MedMeanK, MaxMedK and MaxMeanK) implemented in STRUCTURESELECTOR (Li and Liu 2018). Structure results were summarized with CLUMPAK (Kopelman et al. 2015). Simulations studies have shown that Structure is particularly robust compared to other clustering methods when dosage information is incomplete, even in the presence of mixed ploidies (Stift et al. 2019).
Results
Microsatellite design and analysis
Selected primer pairs, microsatellite motifs, and size ranges are reported in Table 1. The 12 newly designed nuclear microsatellite primer pairs amplified all samples and the resulting loci were variable with some exceptions. In particular, locus Opat_38 was not variable for the Italian O. patens but showed at least two alleles (Table 2) in the Algerian populations, and it did not show any amplification product in the samples from the Canary Islands. Locus Opat_39, with multiple alleles in O. patens from Italy and Algeria (Table 2), was not variable for samples from the Canary Islands.
The number of alleles as inferred by the maximum number of peaks in the electropherograms was four in all populations. Every genotyped individual had at least one locus displaying three peaks, except GC8 in the Canary Islands, which exhibited two peaks at every locus (Table 2). Therefore, we did not find strong evidence of populations with mixed ploidy. The analysis in polysat showed that polysomic inheritance prevailed at all loci, with one exception represented by locus Opat_36 in the Algerian samples (Fig. 2). Consequently, data were treated as for autopolyploid species in the subsequent analyses.
To verify the transferability of these microsatellite loci, we tested the primers in other Orchis taxa (Table 3). Opat_27, Opat_32, Opat_34, Opat_36 and Opat_38 did not amplify O. provincialis samples (with locus Opat_38 being variable only in Algerian O. patens). All the primers were transferable to O. × fallax, the natural hybrid between O. patens and O. provincialis, with the only exception of Opat_13 and Opat_38 that showed no variability (they were not variable in O. provincialis and O. patens, respectively). Six out of 12 primer pairs successfully amplified and showed variability also in O. mascula subsp. mascula and O. mascula subsp. ichnusae samples. Three primer pairs amplified and showed variability also in O. spitzelii. Furthermore, Opat_13 amplified and was variable also in four ‘anthropomorphic’ species: O. anthropophora, O. simia, O. militaris and O. purpurea (Bersweden et al. in rev.).
Genetic diversity and structure
Genetic diversity parameters for each population and locality are summarized in Tables 2 and 4. The results indicate that genetic diversity indices are similar across all populations and localities, except Tenerife, which shows slightly lower heterozygosities and number of alleles (Table 4). The number of effective alleles (Eff_Num) per population varied between 3.2 (in Italy, averaged over 11 polymorphic loci) and 4.4 (in the Canary Islands, averaged over ten polymorphic loci). The lowest Ho was found in the Canary Islands populations (0.49, reflecting the low HO observed in Tenerife; Table 4) and the highest in the Italian populations (0.69); HS ranged between 0.64 in Italy and 0.72 in the Canary Islands (Table 2). Negative values of GIS were found in all populations, possibly reflecting the tetraploid status of the species; however, locus-specific high GIS were found for Opat_20 and Opat_29, possibly indicating null alleles or dropout (i.e., alleles that do not produce amplification by PCR). The highest selfing rate, as computed in SPAGeDI was found in the Algerian locality OPAZ; absence of selfing was estimated in Italian and Canarian localities (Table 4).
The hierarchical AMOVA (Table 5) showed most of the variation at the individual level, which is expected when using polymorphic (and polyploid) microsatellite loci. Moreover, differentiation among groups was the highest for the group formed respectively by Algerian/Italian populations and the Canary Islands populations (Rho_ct: 0.421) and the lowest when one of the groups included Italian and Canarian populations (Rho_ct: 0.162). In parallel, the matrix of pairwise Rho-values indicated strong differentiation among populations in Italy, Algeria and the Canary Islands and also among the individuals collected in different islands of the archipelago (Fig. 3). When performing AMOVA on the Italian subpopulations, for which we had more localities and individuals (i.e., 45), the results (Table 6) pointed to higher levels of variation within (89.8%) rather than between the subpopulations (10.2%).
The neighbor-joining tree based on the square matrix of genotype dissimilarities obtained with polysat indicated three clear clusters (Fig. 4). In particular, the cluster including all the samples from the Canary Islands (light blue) is divergent from the other two. The same pattern was observed in the principal component analysis (PCA) based on Bruvo distances (Fig. 5).
Neighbor-joining tree of Orchis patens s.l. based on a square dissimilarity matrix based on the 12 microsatellite loci. There are three clear clusters consistent with sampling countries. A “canariensis” cluster (TN, GC and LG samples; light blue), an Italian cluster (OP samples; blue) and an Algerian cluster (OPAZ and OPSA samples; gold)
Principal component analysis (PCA) of Orchis patens s.l. based on Bruvo distances among individuals, based on the 12 microsatellite loci. The three major groups are consistent with the Neighbor-joining tree, but it is possible to observe further diversification within populations. In particular, African samples (gold) seem to cluster in two minor groups (samples were collected in two different locations in Algeria). Canary Islands—light blue; Italy—blue; Algeria—gold)
DeltaK values computed on the output of the Structure analysis indicated a strong signal for K = 2 (Supplementary information Fig. S1), whereas the other estimators (MedMedK, MedMeanK, MaxMedK and MaxMeanK) indicated three clusters (Supplementary information Fig. S2). The clusters underlined, once again, a strong genetic differentiation between O. patens s.s. and O. canariensis) (Fig. 6).
a Structure clustering results for 94 samples of Orchis patens s.l. (45 samples from Italy, 20 from Algeria and 29 samples from Canary Islands) based on the variation at 12 microsatellite loci. Barplots are reported for K = 2 (top) and K = 3 (bottom), as determined using the Evanno method (Fig. S1) and the Puechmaille’s estimators (Fig. S2); b Structure barplots of genetic differentiation in O. patens s.s. for all the Italian and Algerian sampling localities included in the present study
The analysis conducted on the Italian and Algerian populations identified four potential clusters (Supplementary information Fig. S3), whereas the analysis conducted on the samples from the Canary Islands identified three clusters (Supplementary information Fig. S6). In both cases, it was possible to observe an important division driven by regionality (Fig. 7) which, in the Canary Islands, are consistent with the three islands where samples were collected. For each of the Algerian and the Italian subsets, two clusters were identified (Supplementary information Figs. S4–S5) (Fig. 7).
Structure barplots of genetic differentiation in Orchis patens s.l. for all the sampling localities included in the present study, based on the variation at 12 microsatellite loci. Each barplot is based on the most likely K value retrieved using the Evanno method and the Puechmaille’s estimators (see Supplementary Figures)
Discussion
Preserving genetic variation is imperative to ensure the existence of any species, especially under anthropogenic and environmental stresses, including climate change (Hewitt 2004; Jump et al. 2009). For this reason, rare species with a narrow distribution or a fragmented habitat, and therefore potentially low genetic diversity, are a priority for studies aimed at investigating genetic diversity to establish appropriate conservation strategies. Orchidaceae are characterized by a diverse range of reproductive strategies (Cozzolino and Widmer 2005; Duffy and Johnson 2017; Schatz et al. 2020), geographic distribution and life histories that are reflected in a variety of patterns in the population genetic structure of different species (Swarts et al. 2009). Clear species delimitations are important for orchids because many of the currently recognized species are of conservation concern and threats continue to increase due to habitat degradation (Cozzolino et al. 2003; Fay 2018). In this study we used 12 newly developed nDNA markers to determine the level of genetic differentiation among populations of the O. patens polyploid group, with a view to resolving taxonomic uncertainties for conservation purposes (Rankou 2011; Swarts et al. 2014; Orsenigo et al. 2016; Calevo et al. 2018; Médail and Baumel 2018).
Genetic structure and taxonomic delimitation
Our analyses of genetic differentiation based on different approaches revealed the same clear clustering of samples (Figs. 3, 4, 5, 6), suggesting that the microsatellite data presented here carry a clear signal of population subdivision. In particular, samples from the Canary Islands are included in a genetic cluster divergent from the Italian and Algerian populations and are, in turn, characterized by genetic subdivision depending on the island of provenance (Figs. 3, 7; Table 5). The failure to amplify a product using Opat_38 in the Canary Island populations further implies genetic divergence between O. canariensis and O. patens s.s. This locus was successfully amplified in the samples from Italy and Algeria. Even though both the Italian and Canarian plants were recently found to share the same mycobiont showing a high level of phylogenetic conservativism in their symbiotic interactions (Calevo et al. 2020), our results support the separation of O. canariensis as a distinct (probably sister) species, as initially described from Tenerife by Lindley (Orchis canariensis, Lindley 1835) based on morphological characters and as recognised by some authors (e.g., Aceto et al. 1999; Bateman et al. 2003) based on phylogenetic analyses. Analysis of the genetic structure based on microsatellites has proved to be informative in revealing cryptic infraspecific structure and subspecies delimitation, for example in the genera Greenwayodendron (Lissambou et al. 2019) and Lophira (Ewédjè et al. 2020).
The genetic structure of O. canariensis, as characterized by the number of subpopulations and the frequencies of different genetic variants (alleles) in each subpopulation (Chakraborty 1993), might be indicative of a high degree of genetic isolation among the subpopulations on the different islands. The strong clustering of Canarian populations (that mirrors the island of provenance) has been observed in other endemic plants of the islands such as Lotus sessilifolius (Yang et al. 2018) and Bethencourtia spp. (Rodríguez-Rodríguez et al. 2018), and the genetic differentiation among island populations may reflect the very beginning of speciation as proposed for the endemic Kleinia neriifolia by Sun and Vargas-Mendoza (2017). This, as suggested by the latter authors, might be expected as the ocean acts as a natural barrier among the islands.
At the same time, there is also some evidence of differentiation between the Italian and Algerian populations of O. patens s.s. (Figs. 3, 4, 5, 6, 7), which provides a genetic basis for the treatment of the Italian populations as a subspecies of Orchis patens. As mentioned for O. canariensis, microsatellite amplifications revealed a subpopulation-specific pattern (Tables 2 and 6) even among samples from Italy and Algeria, which may suggest that mutations in the priming sites are consistent with intraspecific divergence. This may be promoted by the tetraploid status of O. patens, in which the two chromosome sets undergo a range of somatic mutations and diverge if gene flow is reduced (Moody et al. 1993; Meirmans and Liu 2018). Morphologically, the Italian populations are characterized by slightly spotted leaves (Fig. 1d), whereas spots are absent in African individuals (Fig. 1e) and were not mentioned in the first description of the species from Algeria (Desfontaines 1799). Further morphological studies should be focused on detecting morphometric differences among the subspecies based on their entire phenotype, in order to strengthen their taxonomic circumscription and to collect more data about functional differences towards their conservation.
Influence of polyploidy on current genetic diversity patterns
Although populations of O. patens have experienced natural and anthropogenic fragmentation, and fruit set is low and affected by hybridization with the sympatric species O. provincialis, the genetic structure found here reflects historical gene flow, especially between Italy and Algeria. Orchis patens may have been relatively common in the past within its narrow range and therefore the genetic effects of recent fragmentation and reduction in population number may yet to be detected. In polyploid species, loss of genetic diversity by drift is slowed down because of the availability of multiple allele copies compared to diploid species (Moody et al. 1993; Meirmans and Liu 2018). In addition, the effect of gene flow is amplified as more gene copies are involved. Consequently, the effects of fragmentation and genetic erosion may be not evident until more generations have elapsed. The levels of heterozygosity we found in O. patens are compatible with that of other polyploid species (Gargiulo et al. 2019b; Hagl et al. 2020), and in particular tetraploid orchid populations, such as Gymnadenia conopsea (HO = 0.77; Gustafsson and Thorén 2001) and the Dactylorhiza majalis complex (HO = 0.77 averaged across tetraploids; Balao et al. 2016). It is also possible that the current distribution of O. patens reflects long-distance dispersal from one of the three areas towards the other two, although our results do not allow us to speculate further on the most likely explanation.
Regarding the origin of O. patens s.l., the group has been considered sister of the O. spitzelii group based on an analysis of the ITS region (Bateman et al. 2003). These similarities have led to the hypothesis that O. patens could have been derived from the latter species as an autotetraploid, but a recent study based on seed micromorphology as a taxonomic tool also highlighted similarities with the O. mascula group (Calevo et al. 2017), suggesting the possibility of an allopolyploid origin. Our microsatellite analysis did not recover a segregation pattern typical of allopolyploid species (allopolyploidy is generally associated with disomic inheritance, at least for some loci). However, current segregation patterns are not always informative of past events, as the inheritance may shift from disomic to polysomic and vice-versa (Stift et al. 2008). However, more analyses are needed to better understand the origin of O. patens and the potential taxa involved in the allopolyploidization (Ramsey and Schemske 1998; Soltis et al. 2010; Barker et al. 2015).
Conclusions and implications for conservation
Combining molecular evidence from our newly developed nuclear microsatellite markers with the geographical distribution of the studied populations, we propose the recognition of O. patens subsp. canariensis as a separate (possibly sister) species, i.e., O. canariensis, and the treatment of the Italian populations of O. patens as a distinct subspecies. Moreover, the primer set for the 12 microsatellite loci with transferability potential represents a useful tool for interspecific research in the genus Orchis. Our study of species delimitation within the O. patens s.l. complex can offer guidelines for further morphological and genetic investigations and conservation studies. Therefore, we recommend the following actions:
-
Orchis canariensis should be assessed for the IUCN red list;
-
the Mediterranean assessment of O. patens (Calevo et al. 2018) should be revised taking into account the taxonomic delimitation suggested here (which will probably lead to a higher category of threat), the evidence of restricted gene flow, narrow range and low number of individuals (Orsenigo et al. 2016);
-
given that the availability of the main symbiotic fungus of O. patens s.l. at global scale seems not to be a limiting factor for their distribution (Calevo et al. 2020), conservation actions aimed at preventing further habitat fragmentation should be taken. In particular, local authorities and agencies in Algeria, Tunisia (where the natural population is already potentially extinct), Italy and the Canary Islands should institute measures to protect remaining individuals from damage and illicit collection.
-
Further non-destructive sampling, even if difficult due to the rarity and low density of the species, may be required to better investigate the level of inbreeding and its contribution to the reproduction of the populations. Once reproduction mechanisms are clarified, strategies which facilitate pollination should be taken into account, also considering the low fruit-set of the species.
Availability of data and material
Representative sequences were deposited in GenBank (Accession Numbers MT799099-MT799110).
References
Aceto S, Caputo P, Cozzolino S, Gaudio L, Moretti A (1999) Phylogeny and evolution of Orchis and allied genera based on ITS DNA variation: morphological gaps and molecular continuity. Mol Phylogenet Evol 13:67–76
Aedo C, Herrero A (2005) Flora Iberica. Vol. 21: Smilacaceae - Orchidaceae. Madrid
Aguilar R, Quesada M, Ashworth L, Herrerias-Diego Y, Lobo J (2008) Genetic consequences of habitat fragmentation in plant populations: susceptible signals in plant traits and methodological approaches. Mol Ecol 17:5177–5188
Allendorf FW, Luikart G (2007) Conservation and the genetics of populations. Blackwell Publishing, Malden
Andrews S (2010) FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Balao F, Tannhäuser M, Lorenzo M, Hedrén M, Paun O (2016) Genetic differentiation and admixture between sibling allopolyploids in the Dactylorhiza majalis complex. Heredity 116:351–361. https://doi.org/10.1038/hdy.2015.98
Barker MS, Arrigo N, Baniaga AE, Li Z, Levin DA (2016) On the relative abundance of autopolyploids and allopolyploids. New Phytol 210:391–398
Barnosky A, Matzke N, Tomiya S et al (2011) Has the Earth’s sixth mass extinction already arrived? Nature 471:51–57. https://doi.org/10.1038/nature09678
Bateman RM, Hollingworth PM, Preston J, Yibo L, Pridgeon AM, Chase MW (2003) Molecular phylogenetics and evolution of Orchidinae and selected Habenariinae (Orchidaceae). Bot J Linn Soc 142:1–40
Bernardos S, García-Barriuso M, León-Arencibia MC, Reyes-Betancort A, González-González R, Padrón M, Amich F (2006) A cytotaxonomic study of three endemic orchids of the Canary Islands. Ann Bot Fennici 43:161–166
Besnard G, Baali-Cherif D (2009) Coexistence of diploids and triploids in a Saharan relict olive: evidence from nuclear microsatellite and flow cytometry analyses. C R Biol 332:1115–1120
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina Sequence Data. Bioinformatics 30:2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Borrell JS, Zohren J, Nichols RA, Buggs RJ (2020) Genomic assessment of local adaptation in dwarf birch to inform assisted gene flow. Evol Appl 13(1):161–175. https://doi.org/10.1111/eva.12883
Brooks TM, Mittermeier RA, Mittermeier CG, Da Fonseca GAB, Rylands AB, Konstant WR, Flick P, Pilgrim J, Oldfield S, Magin G, Hilton-Taylor C (2002) Habitat loss and extinction in the hotspots of biodiversity. Conserv Biol 16:909–923
Bruvo R, Michiels NK, D’Souza TG, Schulenburg H (2004) A simple method for the calculation of microsatellite genotype distances irrespective of ploidy level. Mol Ecol 13:2101–2106
Calevo J, Giovannini A, Cornara L, Peccenini S, Monroy F (2017) Orchis patens Desf.: seed morphology of an endangered Mediterranean orchid. Plant Biosyst 151:770–774. https://doi.org/10.1080/11263504.2017.1297335
Calevo J, Montagnani C, Véla E (2018) Orchis patens. The IUCN Red List of Threatened Species 2018:e.T175961A84665334. https://doi.org/10.2305/IUCN.UK.2018-2.RLTS.T175961A84665334.en
Calevo J, Voyron S, Ercole E, Girlanda M (2020) Is the distribution of two rare Orchis Sister species limited by their main mycobiont? Diversity 12:262. https://doi.org/10.3390/d12070262
Chakraborty R (1993) Analysis of genetic structure of populations: meaning, methods, and implications. In: Majumder PP (eds) Human population genetics. Springer, Boston
Chase MW, Cameron KM, Barrett RL, Freudenstein JV (2003) DNA data and Orchidaceae systematics: a new phylogenetic classification. In: Dixon KW, Kell SP, Barrett RL, Cribb PJ (eds) Orchid conservation. Natural History Publications, Kota Kinabalu, pp 69–89
Chase MW, Cameron KM, Freudenstein JV, Pridgeon AM, Salazar G, van den Berg C, Schuiteman A (2015) An updated classification of Orchidaceae. Bot J Linn Soc 177(2):151–174. https://doi.org/10.1111/boj.12234
Clark LV, Jasieniuk M (2011) POLYSAT: an R package for polyploid microsatellite analysis. Mol Ecol Resour 11:562–566
Clark LV, Schreier AD (2017) Resolving microsatellite genotype ambiguity in populations of allopolyploid and diploidized autopolyploid organisms using negative correlations between allelic variables. Mol Ecol Resour 17:1090–1103
Cozzolino S, Noce ME, Musacchio A, Widmer A (2003) Variation at a chloroplast minisatellite locus reveals the signature of habitat fragmentation and genetic bottlenecks in the rare orchid Anacamptis palustris (Orchidaceae). Am J Bot 90:1681–1687. https://doi.org/10.3732/ajb.90.12.1681
Cozzolino S, Widmer A (2005) Orchid diversity: an evolutionary consequence of deception? Trends Ecol Evol 20:487–494
Desfontaines RL (1799) Flora Atlantica, sive Historia plantarum, quae in Atlante, agro Tunetano et Algeriensi crescunt, Parigi, L.G. Desgranges
Dodsworth S (2015) Genome skimming for next-generation biodiversity analysis. Trends Plant Sci 20:525–527
Donkpegan AS, Doucet JL, Dainou K, Hardy OJ (2015) Microsatellite development and flow cytometry in the African tree genus Afzelia (Fabaceae, Caesalpinioideae) reveal a polyploid complex. Appl Plant Sci 3(1):1400097
Duffy KJ, Johnson SD (2017) Specialized mutualisms may constrain the geographical distribution of flowering plants. Proc R Soc B 284:20171841
Dufresne F, Stift M, Vergilino R, Mable BK (2014) Recent progress and challenges in population genetics of polyploid organisms: an overview of current state-of-the-art molecular and statistical tools. Mol Ecol 23:40–69
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Ewédjè E-EBK, Jansen S, Koffi GK, Staquet A, Pineiro R, Esaba RA, Obiang NLE, Daïnou K, Biwolé AB, Doucet JL, Hardy OJ (2020) Species delimitation in the African tree genus Lophira (Ochnaceae) reveals cryptic genetic variation. Conserv Genet. https://doi.org/10.1007/s10592-020-01265-7
Fahrig L (2003) Effects of habitat fragmentation on biodiversity. Ann Rev Ecol Evol Syst 34:487–515
Faircloth BC (2008) MSATCOMMANDER: Detection of microsatellite repeat arrays and automated, locus-specific primer design. Mol Ecol Resour 8:92–94
Falush D, Stephens M, Pritchard JK (2007) Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes 7:574–578
Fay MF (2018) Orchid conservation: how can we meet the challenges in the twenty-first century? Bot Stud 59:16
Fay MF (2020) Orchids on the IUCN Global Red List. Orchid Specialist Group Newsletter 2020 (2):2–4. https://www.iucn.org/sites/dev/files/content/documents/osg_newsletter_sept_final.pdf
Fay MF, Gargiulo R, Viruel J (2019) The present and future for population genetics, species boundaries, biogeography and conservation. Bot J Linn Soc 191:299–304
Frankham R, Ballou JD, Briscoe DA (2002) Introduction to conservation genetics. Cambridge University Press, Cambridge, UK
Gargiulo R, Pironon S, Zheleznaya E, Sanchez MD, Balázs ZR, Podar D, Wilkinson T, Jäkäläniemi A, Kull T, Väre H, Fay MF (2019a) Phylogeography and post-glacial dynamics in the clonal-sexual orchid Cypripedium calceolus L. J Biogeogr 46:526–538. https://doi.org/10.1111/jbi.13528
Gargiulo R, Worswick G, Arnold C, Pike LJ, Cowan RS, Hardwick KA, Chapman T, Fay MF (2019) Conservation of the threatened species, Pulsatilla vulgaris Mill. (Pasqueflower), is aided by reproductive system and polyploidy. J Hered 110(5):618–628. https://doi.org/10.1093/jhered/esz035
Garner BA, Hoban S, Luikart G (2020) IUCN Red List and the value of integrating genetics. Conserv Genet 21:795–801. https://doi.org/10.1007/s10592-020-01301-6
Gompert Z, Mock KE (2017) Detection of individual ploidy levels with genotyping-by-sequencing (GBS) analysis. Mol Ecol Resour 17:1156–1167
Gustafsson S, Thorén PA (2001) Microsatellite loci in Gymnadenia conopsea, the fragrant orchid. Mol Ecol Notes 1:81–82
Hagl PA, Gargiulo R, Fay MF, Solofondranohatra C, Salmona J, Suescun U et al (2020) Geographical structure of genetic diversity in Loudetia simplex (Poaceae) in Madagascar and South Africa. Bot J Linn Soc. https://doi.org/10.1093/botlinnean/boaa098
Hamrick JL, Godt MJW (1996) Conservation genetics of endemic plant species. In: Avise JC, Hamrick JL (eds) Conservation genetics. Chapman and Hall, New York, pp 281–304
Hardy OJ (2016) Population genetics of autopolyploids under a mixed mating model and the estimation of selfing rate. Mol Ecol Resour 16(1):103–117
Hautzinger L (1978) Genus “Orchis” L. (Orchidaceae): Sectio “Robustocalcare” Hautzinger. Ann Nat Mus Wien 81:31–73
Hardy OJ, Vekemans X (2002) SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes 2:618–620
Henle K, Lindenmayer DB, Margules CR, Saunders DA, Wissel C (2004) Species survival in fragmented landscapes: where are we now? Biodivers Conserv 13:1–8
Hewitt GM (2004) Genetic consequences of climatic changes in the Quaternary. Philos Trans R Soc Lond B 359:183–195
Hoban S, Bruford M, Jackson JDU, Lopes-Fernandes M, Heuertz M, Hohenlohe PA et al (2020) Genetic diversity targets and indicators in the CBD post-2020 Global Biodiversity Framework must be improved. Biol Conserv 248:108654
Honnay O, Jacquemyn H (2007) Susceptibility of common and rare plant species to the genetic consequences of habitat fragmentation. Conserv Biol 21:823–831
IUCN (2020) The IUCN Red List of threatened species. Available at: https://www.iucnredlist.org/
Jump AS, Marchant R, Penuelas J (2009) Environmental change and the option value of genetic diversity. Trends Plant Sci 14:51–58
Kretzchmar H, Eccarius W, Dietrich H (2007) The Orchid Genera Anacamptis, Orchis, Neotinea. EchinoMedia, Bürgel
Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA (2015) CLUMPAK: a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Resour 15(5):1179–1191. https://doi.org/10.1111/1755-0998.12387
Laikre L, Hoban S, Bruford MW, Segelbacher G, Allendorf FW, Gajardo G et al (2020) Post-2020 goals overlook genetic diversity. Science 367:1083–1085
Landoni B, Viruel J, Gómez R, Allaby RG, Brennan AC, Picó FX, Pérez-Barrales R (2020) Microsatellite marker development in the crop wild relative Linum bienne using genome skimming. App Plant Sci 8(5):e11349
Leimu R, Mutikainen P, Koricheva J, Fischer M (2006) How general are positive relationships between plant population size, fitness, and genetic variation? J Ecol 94:942–952
Li YL, Liu JX (2018) StructureSelector: a web-based software to select and visualize the optimal number of clusters using multiple methods. Mol Ecol Resour 18:176–177. https://doi.org/10.1111/1755-0998.12719
Lindley J (1835) Genera and species of orchidaceous plants. 263 London, Ridgways Picadilly
Lissambou BJ, Couvreur TL, Atteke C et al (2019) Species delimitation in the genus Greenwayodendron based on morphological and genetic markers reveals new species. Taxon 68(3):442–454
Médail F, Baumel A (2018) Using phylogeography to define conservation priorities: the case of narrow endemic plants in the Mediterranean Basin hotspot. Biol Conserv 224:258–266
Meirmans PG (2012) AMOVA-based clustering of population genetic data. J Hered 103(5):744–750
Meirmans PG (2020) genodive version 3.0: easy‐to‐use software for the analysis of genetic data of diploids and polyploids. Mol Ecol Resour 20:1126–1131
Meirmans PG, Van Tienderen PH (2004) GENOTYPE and GENODIVE: two programs for the analysis of genetic diversity of asexual organisms. Mol Ecol Notes 4:792–794
Meirmans PG, Liu S (2018) Analysis of Molecular Variance (AMOVA) for autopolyploids. Front Ecol Evol 6:66
Meirmans PG, Liu S, Van Tienderen PH (2018) The analysis of polyploid genetic data. J Hered 109:283–296
Mkare TK, van Vuuren BJ, Teske PR (2017) Conservation implications of significant population differentiation in an endangered estuarine seahorse. Biodivers Conserv 26:1275–1293. https://doi.org/10.1007/s10531-017-1300-5
Mrkvicka A (1992) Liste der Chromosomenzahlen europäischer Orchideen. Mitteilungsbl Arbeitskreis Heimische Orchid Baden-Württemberg 24(1):669–678
Moody ME, Mueller LD, Soltis DE (1993) Genetic variation and random drift in autotetraploid populations. Genetics 134:649–657
Nemorin A, David J, Maledon E et al (2013) Microsatellite and flow cytometry analysis to help understand the origin of Dioscorea alata polyploids. Ann Bot 112:811–819
Orsenigo S, Bacchetta G, Calevo J, Castello M, Cogoni D, Gennai M et al (2016) Global and regional IUCN red list assessments: 1. Italian Bot 1:61–85. https://doi.org/10.3897/italianbotanist.1.8647
Pandey M, Richards M, Sharma J (2015) Microsatellite-based genetic diversity patterns in disjunct populations of a rare orchid. Genetica 143:693–704. https://doi.org/10.1007/s10709-015-9867-9
Petit RJ, Mousadik AE, Pons O (1998) Identifying populations for conservation on the basis of genetic markers. Conserv Biol 12:844–855
Pridgeon AM, Cribb PJ, Chase MW, Rasmussen FN (eds.) (2001) Genera Orchidacearum, vol. 2. Oxford University Press, Oxford
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Puechmaille SJ (2016) The program structure does not reliably recover the correct population structure when sampling is uneven: subsampling and new estimators alleviate the problem. Mol Ecol Resour 16:608–627. https://doi.org/10.1111/1755-0998.12512
Ramsey J, Schemske DW (1998) Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu Rev Ecol Evol Syst 29:467–501
Rankou H (2011) Orchis patens. The IUCN Red List of Threatened Species
Rodríguez-Rodríguez P, Pérez de Paz PL, Sosa PA (2018) Species delimitation and conservation genetics of the Canarian endemic Bethencourtia (Asteraceae). Genetica 146:199–210. https://doi.org/10.1007/s10709-018-0013-3
Ronfort JL, Jenczewski E, Bataillon T, Rousset F (1998) Analysis of population structure in autotetraploid species. Genetics 150:921–930
Schatz B, Genoud D, Claessens J, Kleynen J (2020) Orchid-pollinator network in Euro-Mediterranean region: what we know, what we think we know, and what remains to be done. Acta Oecol. https://doi.org/10.1016/j.actao.2020.103605
Schwartz MK, Luikart G, Waples RS (2007) Genetic monitoring as a promising tool for conservation and management. Trends Ecol Evol 22:25–33
Spielman D, Brook BW, Frankham R (2004) Most species are not driven to extinction before genetic factors impact them. Proc Natl Acad Sci USA 101:15261–15264
Soltis DE, Buggs RJA, Doyle JJ, Soltis PS (2010) What we still don’t know about polyploidy. Taxon 59:1387–1403
Stark C, Michalski SG, Babik W, Winterfeld G, Durka W (2011) Strong genetic differentiation between Gymnadenia conopsea and G. densiflora despite morphological similarity. Plant Syst Evol 293:213–226. https://doi.org/10.1007/s00606-011-0439-x
Stift M, Berenos C, Kuperus P, Van Tienderen PH (2008) Segregation models for disomic, tetrasomic and intermediate inheritance in tetraploids: a general procedure applied to Rorippa (yellow cress) microsatellite data. Genetics 179:2113–2123
Stift M, Kolář F, Meirmans PG (2019) Structure is more robust than other clustering methods in simulated mixed-ploidy populations. Heredity 123:429–441. https://doi.org/10.1038/s41437-019-0247-6
Straub SC, Parks M, Weitemier K et al (2012) Navigating the tip of the genomic iceberg: Next-generation sequencing for plant systematics. Am J Bot 99(2):349–364
Sun Y, Vargas-Mendoza CF (2017) Population structure, genetic diversity, and evolutionary history of Kleinia neriifolia (Asteraceae) on the Canary Islands. Front Plant Sci 8:1180. https://doi.org/10.3389/fpls.2017.01180
Swarts ND, Dixon KW (2009) Terrestrial orchid conservation in the age of extinction. Ann Bot 104(3):543–556. https://doi.org/10.1093/aob/mcp025
Swarts ND, Sinclair EA, Krauss SL, Dixon KW (2009) Genetic diversity in fragmented populations of the critically endangered spider orchid Caladenia huegelii: implications for conservation. Conserv Genet 10:1199–2120
Swarts ND, Clements MA, Bower CC, Miller JT (2014) Defining conservation units in a complex of morphologically similar, sexually deceptive, highly endangered orchids. Biol Conserv 174:55–64
Teixeira H, Rodrıguez-Echeverría S, Nabais C (2014) Genetic Diversity and Differentiation of Juniperus thurifera in Spain and Morocco as Determined by SSR. PLoS ONE 9(2):e88996
Väli Ü, Dombrovski V, Dzmitranok M, Maciorowski G, Meyburg BU (2019) High genetic diversity and low differentiation retained in the European fragmented and declining Greater Spotted Eagle (Clanga clanga) population. Sci Rep 9:3064. https://doi.org/10.1038/s41598-019-39187-1
Vellend M, Geber MA (2005) Connections between species diversity and genetic diversity. Ecol Lett 8:767–781
Viruel J, Haguenauer A, Juin M et al (2018) Advances in genotyping microsatellite markers through sequencing and consequences of scoring methods for Ceratonia siliqua (Leguminosae). Appl Plant Sci 6(12):e1201. https://doi.org/10.1002/aps3.1201
Viruel J, Conejero M, Hidalgo O et al (2019) A Target capture-based method to estimate ploidy from herbarium specimens. Front Plant Sci 10:937. https://doi.org/10.3389/fpls.2019.00937.PMID:31396248;PMCID:PMC6667659
Volkmann L, Martyn I, Moulton V, Spillner A, Mooers AO (2014) Prioritizing populations for conservation using phylogenetic networks. PLoS ONE 9(2):e88945. https://doi.org/10.1371/journal.pone.0088945
WCSP (2020) World Checklist of Selected Plant Families. Facilitated by the Royal Botanic Gardens, Kew. Published on the Internet; http://wcsp.science.kew.org/ Retrieved on 13 March 2020
Yang JY, Ojeda DI, Santos-Guerra A, Molina RJ, Caujapé-Castells J, Cronk Q (2018) Population differentiation in relation to conservation: nuclear microsatellite variation in the Canary Island endemic Lotus sessilifolius (Fabaceae). Conserv Genet Resour 10(2):219–227
Young A, Boyle T, Brown T (1996) The population genetic consequences of habitat fragmentation for plants. Trends Ecol Evol 11:413–418
Acknowledgements
Authors are truly grateful to Robyn Cowan, László Csiba and Gabriele Rizzuto for technical support, and to Dr. Pierluigi Cortis, Dr. Errol Vela, Dr. Selim Amine, Dr. Giampiero Ciaschetti-National Park of Majella, and Dr. Ángel Fernández López-National Park of Garajonay for logistic and sampling support. We thank Prof. Richard Bateman and one anonymous reviewer for providing insightful comments which improved the quality of our manuscript.
Funding
This research was conducted within the framework of the PhD program of Jacopo Calevo funded by the Italian Ministry of Education and Research (MIUR).
Author information
Authors and Affiliations
Contributions
JC, CGM, KR and LBou. collected samples; JC, MFF, RG and JV designed research; JC and LBers. performed research; JC and RG analyzed data, JC wrote the paper. All authors reviewed and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Daniel Sanchez Mata.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Calevo, J., Gargiulo, R., Bersweden, L. et al. Molecular evidence of species- and subspecies-level distinctions in the rare Orchis patens s.l. and implications for conservation. Biodivers Conserv 30, 1293–1314 (2021). https://doi.org/10.1007/s10531-021-02142-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10531-021-02142-6