Abstract
Naturalized species of alien plants and animals comprise < 3% of biodiversity recorded in Norway but have had major impacts on natural ecosystems through displacement of native species. Encroachment of alien species has been especially problematic for coastal sites close to transport facilities and urban areas with high density housing. The goal of our field project was to design and test a surveillance program for early detection of alien species of vascular plants and terrestrial insects at the first phase of establishment in natural areas. In our 3-year project (2018–2020), we sampled 60 study plots in three counties in the Oslofjord region of southern Norway. Study plots (6.25 ha) were selected by two criteria: manual selection based on expert opinion (27 plots) or by random selection based on weights from a hotspot analysis of occurrence of alien species (33 plots). Vascular plants were surveyed by two experienced botanists who found a total of 239 alien species of vascular plants in 95 rounds of surveys. Insects and other invertebrates were captured with a single Malaise trap per site, with 3–4 rounds of repeated sampling. We used DNA-metabarcoding to identify invertebrates based on DNA extractions from crushed insects or from the preservative media. Over 3500 invertebrate taxa were detected in 255 rounds of sampling. We recorded 20 alien species of known risk, and 115 species that were new to Norway, including several ‘doorknocker’ species identified by previous risk assessments. We modeled the probabilities of occupancy (ψ) and detection (p) with occupancy models with repeated visits by multiple observers (vascular plants) or multiple rounds of sampling (insects). The two probabilities covaried with risk category for alien organisms and both were low for species categorized as no known or low risk (range = 0.052–0.326) but were higher for species categorized as severe risk (range = 0.318–0.651). Selecting sites at random or manually did not improve the probability of finding novel alien species, but occupancy had a weak positive relationship with housing density for some categories of alien plants and insects. We used our empirical estimates to test alternative sampling designs that would minimize the combined variance of occupancy and detection (A-optimality criterion). Sampling designs with 8–10 visits per site were best for surveillance of new alien species if the probabilities of occupancy and detection were both low, and provided low conditional probabilities of site occupancy (\(\hat{\psi }_{condl}\) ≤ 0.032) and a high probabilities of cumulative detection (\(\hat{p}*\) ≥ 0.943). Our field results demonstrate that early detection is feasible as a key component of a national surveillance program based on early detection and rapid response.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Prevention of establishment and secondary spread of invasive species requires information on the main pathways of introduction for alien organisms and their propagules (Lodge et al. 2006; Meyerson and Mooney 2007; Hulme 2009; Chapman et al. 2017), their species composition and dispersal capability (Fremstad and Elven 1997), and a risk assessment of the potential threat from newly discovered alien species (Reaser et al. 2020; Sandvik et al. 2020). In the case of vascular plants and insects, introductions are primarily associated with horticultural and agricultural production, and secondarily by shipment of soil, timber or other construction materials, or by deliberate planting of fast-growing species to stabilize soils and reduce erosion (Lodge et al. 2006; Olsen et al. 2017). International trade of live plants that are produced in nurseries and then distributed for planting in domestic gardens can be a source of invasive organisms associated with the soils or plant tissues, or the ornamental plants themselves can become invasive (Reichard and White 2001; Qvenild et al. 2014; Westergaard et al. 2018). Secondary spread of plants and insects out from suburban gardens into natural areas can occur through active or passive dispersal of seeds or vegetative propagules, or by dumping of garden refuse (Fremstad and Elven 1997; Ødegaard and Tømmerås 2000). Spread can also occur via long distance dispersal or ‘jumps’ from newly established populations if invasive species or their vectors have high dispersal capability (Bennett et al. 2011; Tavecchia et al. 2017), which may be facilitated by new environmental conditions associated with global climate change (Alpert 2006; Bebber et al. 2013; Bellard et al. 2018). Risk assessments of threat are then based on multiple criteria, including the life-history traits that influence invasion potential, expansion speed and colonized area, and ecological impacts on native species and vulnerable ecosystems (Sandvik et al. 2020). Invasive plants and insects often have high reproductive rates and short generation times that favor rapid population growth, vegetative growth or parthenogenesis that favor spread of clones, and natural defenses that reduce losses to herbivory or predation (Sakai et al. 2001; Sutherland 2004; Milbau and Stout 2008).
Norway is a northern country with a rugged terrain that includes deep coastal fjords, islands, and mountainous areas. The terrestrial habitats are resistant to invasive species because the climate is cold with a short growing season, and only 3.8% of the land base is arable farmland with the rest in boreal forests, wetlands and alpine habitats (Wasof et al. 2015; Bryn et al. 2018). The Norwegian Biodiversity Information Centre (Artsdatabanken in Norwegian) maintains national databases of native and alien species (2015 Species Nomenclature Database, Artsdatabanken 2015), and coordinates risk assessments for alien species at regular intervals (2018 Norwegian Alien Species List, Artsdatabanken 2020; Sandvik et al. 2020). Baseline data on diversity of plants and insects in Norway have been collected by museum systematists, by amateur naturalists who register records in the Species Observations system (Artsobservasjoner), and in national survey programs (Olsen et al. 2017; Åström et al. 2022). Taxonomic coverage is good for well-studied groups of organisms (beetles, butterflies, and vascular plants: Fremstad and Elven 1997; Enderstøl and Økland 2019), but new discoveries can still be made for poorly known groups. A total of 3142 alien species have been registered in Norway (Artsdatabanken 2020), of which at least 1039 species (33.1%) are considered naturalized (Sandvik et al. 2019). Overall, naturalized alien species comprise 2.8% of the 36,872 species of plants and animals recorded in Norway. The percentage of aliens varies among taxonomic groups, ranging from 0.7% among freshwater and terrestrial invertebrates (23,120 species), 1.9% of vertebrates (1266 species), and up to 21.6% of vascular plants (3405 species). Naturalized aliens are not evenly distributed in Norway, with higher species numbers in counties along the southern coast, which includes low elevation habitats with higher mean temperatures and precipitation, and also urban areas with more infrastructure and higher human population densities (Olsen et al. 2017). Established species of alien plants and insects have negatively impacted natural ecosystems in Norway through displacement of native species (Blaalid and Often 2019; Endrestøl and Økland 2019; Artsdatabanken 2020), especially highly invasive species such as Japanese knotweed (Reynoutria japonica), Canadian goldenrod (Solidago canadensis), garden lupin (Lupinus polyphyllus), Asian lady beetles (Harmonia axyridis), and bark beetles (Ips amitinus).
A useful framework for management of invasive species is Early Detection and Rapid Response (EDRR, Lodge et al. 2006; Lyal and Miller 2020; Reaser et al. 2020). The process of ‘early detection’ is based on surveys to detect alien species upon arrival or early stages of establishment and before they start to cause harm. A ‘rapid response’ anticipates that measures are more likely to be cost-effective if populations are small or localized and more successful for control of spread or eradication (Simberloff 2003). A comprehensive surveillance program requires stratified sampling of important habitats (Olsen et al. 2017), effective tools for rapid identification of unknown organisms (Darling and Blum 2007; Lyal and Miller 2020), and sampling designs that control for imperfect detection of alien species (Tavecchia et al. 2017; Davis et al. 2018; Fossøy et al. 2020). Surveillance for alien invertebrates can be particularly challenging because sorting and morphological identification from bulk samples is time-consuming and costly (Karlsson et al. 2020), but molecular methods based on DNA-metabarcoding provide a new tool for rapid biodiversity assessments (Yu et al. 2012; Comtet et al. 2015; Wang et al. 2018). Similarly, occupancy models improve upon traditional diversity estimators for alien species because they allow the detection process to be modeled as a function of different covariates (Iknayan et al. 2014). The objectives of our field project were fivefold: (i) to design a systematic surveillance program for detection of alien species of vascular plants and invertebrates at an early stage of establishment in Norway, (ii) to compare site selection procedures that might increase the chance of locating novel alien species in a region, (iii) to evaluate DNA-metabarcoding as a new technique for rapid processing and identification of invertebrates from field samples, (iv) to test occupancy models for estimating the probabilities of occupancy and detection for alien species in different categories of potential risk, and (v) to explore sampling designs for the optimum number of visits per site to achieve high confidence that an alien species would not be overlooked but would be successfully detected in a national surveillance program.
Methods
Selection of study plots
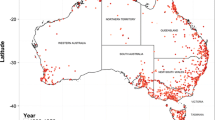
Our project was designed to produce pilot data for a national surveillance program for alien plants and invertebrates in Norway. Our sampling frame was centered on the Oslofjord region of southern Norway and included field sites in the three adjacent counties of Vestfold and Telemark, Viken, and Oslo (Fig. 1). We focused on the Oslofjord region because our preliminary surveys and modeling showed this area has the highest diversity and abundance of alien species in Norway (Olsen et al. 2017; Sandvik et al. 2019), and because our field personnel with expertise in botany and entomology were located in Oslo. For site selection, we used a national grid for Norway with a cell area of 6.25 ha (250 × 250 m) from Statistics Norway. Spatial statistics for each cell were compiled from public GIS layers for average annual temperatures and rainfall (Meteorological Institute), land cover and housing density (AR5 classification system, Norwegian Institute of Bioeconomy Research), and human population numbers (Statistics Norway). We sampled a total of 60 sites, including 15 sites in 2018, 20 sites in 2019, and 25 sites in 2020 (including 4 sites previously sampled in 2019).
Locations of study plots for surveying alien plants and insects in the Oslofjord region of Norway (inset) during 2018 (red, n = 15), 2019 (blue, n = 20), and 2020 (green, n = 25). Sites with solid squares were selected at random from sites that met predefined criteria whereas sites with white dots were manually selected by observers
We used two criteria for selection of our 6.25 ha study plots: manual selection based on expert opinion for the probability of finding new invasive species and random selection based on a set of predefined criteria (Fig. 2). A total of 27 plots were selected manually (7 in 2018 and 10 each in 2019 and 2020) because the plots were located at major transport hubs for shipping or receiving of goods and materials, or because they included recycling depots for garden refuse. Four manually selected sites were surveyed in both 2019 and 2020 but we treated the repeated surveys as independent for our analyses. An additional 33 study plots were selected at random (8 in 2018, 10 in 2019, 15 in 2020) from sites that met four selection criteria: (i) a housing density of at least 8–12 detached houses, (ii) a population density between 30 and 125 residents, (iii) site locations within 100 m of a natural forest area, and (iv) a probability of selection that was weighted by the predicted proportion of alien vascular plants from a recent ‘hotspot’ analysis by Olsen et al. (2017). We generated a set of random sites and then matched a subset of sites by relative proximity to the manually selected sites. Sites were matched to reduce possible differences in environmental conditions between manual and random sites, and to help with logistics of field sampling in a large region. Last, we used the manual and random selection criteria to identify an additional set of backup sites. We conducted a preliminary inspection of all sites before field sampling began to confirm site conditions, to request site access from private landowners, and to arrange necessary permits for sample sites located in nature reserves. If we were unable to arrange access to a selected site, we substituted one of our alternate sites. In a few cases, the optimal site for the Malaise trap was outside of the selected study plot. In these cases, we adjusted the study plot so that it matched the 6.25 ha cell that included the location of the Malaise trap.
Examples of study plots surveyed for alien plants and insects in the Oslofjord region of Norway, 2018–2020. Plots were selected manually as sites with shipping containers or garden refuse (left), or at random based on predefined criteria for housing and population density (right). Plants were surveyed with two rounds of sampling by different observers in a 250 × 250 m study plot (red quadrat) whereas insects were sampled with 3–4 rounds of sampling with a Malaise trap (orange dot)
Field sampling of vascular plants
Two rounds of plant surveys were conducted in the 1-month period between mid-August and mid-September by two experienced botanists who were familiar with the flora of the Oslofjord region (A. Often and H. Hegre). We used a double-observer approach and the two observers conducted independent plant surveys at the study plots. Both observers sampled all 15 sites in 2018. To increase the number of plots visited in 2019 and 2020, 10 plots were sampled by both observers each year, and the remaining plots were sampled by a single observer (5 plots each in 2019, 15 plots by a single observer in 2020). Repeated sampling for a subset of study sites allowed us to use occupancy models to calculate probabilities of detection and occurrence for alien plant species. The procedure for searching plots was adjusted and refined between the three sampling years. In 2018, the study plots were subdivided into four transects offset at 45° angles. Observers were asked to search a 10 m strip along each transect, then spend ca. 30 min searching the remainder of the study plot, with a maximum time limit of two hours per plot. Fixed transects proved to be impractical in residential and industrial areas, and search time was dependent on the habitat complexity of the study plot. In 2019 and 2020, observers used a random walk to search the entire plot, with a maximum time limit of five hours per plot.
Observers recorded native and alien species of vascular plants encountered in natural areas, roadsides, ditches, and other suitable habitats. Cultivated plants were not recorded if they were located in the yards of private homes, park flower beds, or in community gardens. Native species of plants were too numerous to survey systematically but observers recorded species presence opportunistically. If the observers encountered an alien species from the Norwegian Alien Species List (Artsdatabanken 2020), they ranked relative abundance on a 6-point scale: (i) < 5 patches with < 10 individuals or shoots, (ii) < 5 patches with > 10 individuals or shoots, (iii) 5–15 patches with < 10 individuals or shoots, (iv) 5–15 occurrences with > 10 individuals or shoots, (v) a scattered presence in the study plot, and (vi) a dominant presence in the study plot. All detections were weighted equally in the occupancy analyses. Alien plants with few previous records in Norway were documented with photographs and with GPS coordinates of the exact location.
Field sampling of invertebrates
Insects and other invertebrates were sampled with a single Malaise trap at each study site in the 1.5-month period between mid-June and late August. Malaise traps are efficient for collecting large numbers of flying and crawling insects from a diverse range of taxa, and also allow specimens to be preserved for later processing with morphological or genetic analyses (Karlsson et al. 2020). We visited study sites to find a suitable location, and set up Malaise traps in open terrain with good exposure to increase capture efficiency. In 2018, Malaise traps were set up on the study plots in the last week of August (week 35), and we completed three rounds of weekly sampling in September (weeks 36–39). In 2018, the collection bottles were filled with a mixture of water, ethanol, and propylene glycol in a 1:1:3 ratio. In 2019 and 2020, Malaise traps were set up on plots in mid-June (week 25), and we completed four rounds of biweekly sampling between mid-July and late August (weeks 29–35). In 2019–2020, the collection bottles were filled with ca. 400 ml of 96% ethanol. After emptying the traps, the preservative fluid was removed with a sieve and insects were stored in 96% ethanol in the freezer for subsequent analyses. Traps were dismantled and removed after the last round of sampling.
Identification of invertebrates by DNA-metabarcoding
We explored different methods of sample preparation and used DNA-metabarcoding procedures for simultaneous identification of multiple taxa of invertebrates in our samples (Yu et al. 2012; Comtet et al. 2015; Wang et al. 2018). Over the course of the 3 sampling years, we tested DNA-metabarcoding of: (i) homogenised insects, (ii) the preservative ethanol from the traps, and (iii) non-destructive lysis of intact insects. In 2018, insects from the malaise traps were crushed into a homogenous mixture prior to isolation of total DNA for metabarcoding. We were concerned that large-bodied or numerically abundant species might contain relatively more copies of target DNA compared to small-bodied or rare species (Elbrecht et al. 2017). Thus, we first processed the samples by sorting out the large-bodied species of harvestmen (Opiliones), butterflies and moths (Lepidoptera), bumblebees and wasps (Hymenoptera), and caddisflies (Trichoptera), but then returned one or more legs from each specimen to the sample prior to homogenisation (after Braukmann et al. 2019). We then stored the rest of the specimens separately to compare results from morphological analyses versus DNA-metabarcoding procedures. For DNA extractions, insect samples were first dried to remove all ethanol. The insects were then crushed and mixed with 10 mL of animal tissue lysis (ATL) buffer (Qiagen) and 1 mL of proteinase-K (> 600 mAU/ml activity) and 15 ceramic beads in FastPrep 50 mL Matrix E tubes (MP Biomedicals). Samples were lysed overnight in a heating chamber before being isolated with a NucleoSpin Plant II Midi kit (Macherey–Nagel).
Homogenisation of the insect samples was highly effective for DNA extractions, but was also a destructive sampling method that precluded confirmation of species identity by inspection of specimen morphology. Starting in 2019, we tested whether DNA extraction from the preservative ethanol in the field samples or direct lysis of the insect mass could provide an effective but comparatively non-destructive method. The preservative ethanol from Malaise trap samples was filtered first through a 0.3 mm mesh filter, followed by a 0.45 µm cellulose nitrate filter (Nalgene Analytical Test Filter Funnels, ThermoFisher Scientific) using a peristaltic pump (Microsart e.jet, Sartorius GmbH) connected to a 3-arm manifold (Pall Corporation). The cellulose-nitrate filters were placed in 2 mL Eppendorf tubes containing 1440 µL ATL buffer (Qiagen) and 160 µL proteinase-K (> 600 mAU/ml activity, Qiagen) and incubated overnight in a warming cabinet at 56 °C. The filters were then discarded and DNA was isolated from the buffer solution using a NucleoSpin Plant II Midi kit (Mache-rey-Nagel) and eluted in 200 µL AE buffer (Qiagen). The remaining insect mass was then returned to 96% ethanol. For a subset of 10 samples, intact specimens of beetles (Coleoptera) and butterflies (Lepidoptera) were sorted out and identified morphologically by experienced entomologists (K. Berggren and O. Hanssen). One to two legs were removed from each specimen and crushed in a 2 mL FastDNA SPIN Kit for Soil (MP Biomedicals), and DNA was then further extracted with a Blood and Tissue Kit (Qiagen). We then took the remaining 70 samples collected in 2019, filtering them to remove the ethanol preservative, and dried them in a warming cabinet at 56 °C. Dried samples were then lysed by suspending them in a solution of ATL buffer and proteinase-K (100 ml ATL:1 ml proteinase-K with > 600 mAU/ml activity) and incubating them overnight at 56 °C. A 200 μL subsample of the buffer solution was then used for DNA isolation with a Blood and Tissue kit (Qiagen). Based on the improved taxon-recovery rates using the non-invasive sampling protocol, all samples from 2020 were analysed using the lysis method.
Samples from all 3 years were subjected to DNA metabarcoding of the COI gene region using the BF3-BR2 primer set, which provides good taxonomic resolution in arthropod metabarcoding (Elbrecht et al. 2019). We used a standard two-step Ilumina protocol to generate our DNA libraries for metabarcoding (Bohmann et al. 2022). An initial PCR reaction included BF3-BR2 primers with ‘overhang adapter’ sequences, and was followed by a second PCR reaction that appended Illumina Nextera indices (Table S1). PCR products purified after each PCR reaction using a 1:1 ratio of magnetic beads (MAG-BIND RXN PURE PLUS) to sample. The samples were then normalized, pooled and sequenced with 2 × 300 bp on an Illumina MiSeq machine at the Genomics Core Facility (GFC) at the Norwegian University of Science and Technology (NTNU).
Bioinformatics
Sequences were demultiplexed on the MiSeq platform, and the forward and reverse primers were removed from each read using cutadapt ver. 1.18 (Martin 2011), requiring a minimum match length of 17 bp, no indels, and < 0.15 expected errors over the primer length. Quality filtering, error correcting, merging and chimera checking were all conducted using the DADA2 ver. 1.18.0 package in R ver. 4.1 (Callahan et al. 2016). All sequences that were < 50 bp, contained ambiguous bases or > 2 expected errors across the entire length were removed from the dataset, and sequences were truncated at the first instance of a base with Phred quality score < 15. Forward and reverse reads were merged with a minimum overlap of 30 bp and the de novo consensus method in DADA2 was used to remove chimeric sequences. A naïve Bayesian classifer, the RDP classifier (Wang et al. 2007) was used to assign taxonomy to the amplicon sequence variants (ASVs) based on a custom database combining the COI Eukaryote reference set ver. 4 (Porter 2017; Porter and Hajibabaei 2018), as well as ~ 4000 additional publicly available reference sequences for insect species in Norway. For successful assignment at a given taxonomic level, we required a minimum of 80% confidence for the placement. We classified identified species as native or alien in Norway by cross-checking detected taxa against the national databases for biodiversity and alien species (Artsdatabanken 2015, 2020). Despite being the best available resources for Norway, we expected that coverage of alien species would likely be incomplete. We detected a number of species that have not previously been reported in Norway and have not yet been considered for risk assessments for the Norwegian alien species list. The new species (NW) were included as their own category for further analyses and were expected to be an unknown mixture of alien species and native taxa from poorly studied groups that have been previously overlooked.
Statistical analyses
Statistical analyses were conducted in an R environment (ver. 4.0, R Core Team 2019). We extracted six explanatory variables for local environmental conditions at the 60 study plots sampled in 2018–2020: percent land cover in housing, roads, forest, open fields, annual precipitation, and population densities. We tested for pairwise correlations among the explanatory variables with the chart.Correlation function of the PerformanceAnalytics package.
Identification methods differed between the two groups of organisms: alien plants were identified to species by botanists, whereas alien insects and other invertebrates were identified by matching DNA sequences to reference databases and by entomologists. We categorized detected species of alien organisms based on risk assessments from the 2018 Norwegian Alien Species List: no known impact (NK), low impact (LO), potentially high impact (PH), high impact (HI), or severe impact (SE, Artsdatabanken 2020; Sandvik et al. 2020). Newly discovered species of invertebrates (NW) had not been previously reported in Norway (Artsdatabaken 2015), but included species already known to be alien in Europe (NOBANIS 2019), ‘door-knocker’ species where risk assessments have predicted establishment in Norway in the next 50 years (Sandvik et al. 2020), and poorly studied taxa which had been detected in neighbouring Sweden (Karlsson et al. 2020). We censored records of species with no risk (NR) that were considered in the risk assessment but did not meet the criteria to be categorized as alien species. To assess the state of knowledge for each alien species detected in our project, we tallied the number of records reported for each species in the national database for the Species Map (Artskart2, Supplementary Tables S2 and S4).
We used single-season occupancy models to model the probability of occurrence while controlling for false absences (MacKenzie et al. 2006). We expected that alien species might have low probabilities of occupancy (ψ) if they were relatively rare, and that they might have low probabilities of detection (p) if they were cryptic or difficult to identify. We then developed encounter histories for each alien species at each of the 60 study plots. We recorded whether each species was detected (1) or not detected (0) in two independent rounds of sampling by the botanists (plants) or during the 3–4 rounds of sampling from the Malaise traps (insects). All sampling was conducted during a relatively short period < 1.5 mos and we assumed that communities were closed to gains or losses due to differences in seasonal phenology. Violations of the closure assumption can lead to overestimates of occupancy but bias can be minimized if the number of sampling occasions are few, which was the case in our study (Rota et al. 2009; Otto et al. 2013). We conducted goodness-of-fit tests to the global model with the parboot function of the unmarked package and calculated the overdispersion factor (\(\hat{c}\)) as the ratio of chi-square statistics from the observed data versus a bootstrap distribution based on 1000 iterations. We then fit single-season occupancy analyses with the occu function, and ranked the candidate models by Akaike’s Information Criterion (AICc) with the aictab function of the AICcmodavg package. If adjustments for overdispersion were necessary, \(\hat{c}\) was included in the parameter count for the model, and model rankings were based on QAICc.
We used our empirical estimates of occupancy (\(\hat{\psi }\)) and detection (\(\hat{p}\)) for alien species of different risk categories to explore alternative sampling designs for a national surveillance program in Norway. In our 3-year study, we sampled 60 sites (S) with 2–4 rounds of sampling per site (K) for a total sampling effort (TS) of 95 site-visits for plants and 225 site-visits for insects. Sampling designs usually favour fewer visits to more sites for rare or conspicuous species, but more visits to fewer sites for common or cryptic species (MacKenzie et al. 2006:168). We considered alternative study designs for a potential project with ca. 50 sites with at least two rounds of sampling in three regions (Bergen, Oslo, and Trondheim) for a total sampling effort of TS = 300 site-visits. The optimum allocation of effort into number of visits per site versus number of sites (TS = S × K) can be based on several different criteria for study design: minimizing the variance of occupancy alone, minimizing the sum of the variances for occupancy and detection (A-optimality), or minimizing the generalized variance (D-optimality, Guillera-Arroita et al. 2010). We opted to use the A-optimality criterion because surveillance programs require precise estimates for both occupancy and detection for alien species, and because preliminary analyses showed that the two optimality criteria gave similar results. We used numerical simulations based on 100,000 iterations to explore model performance for different combinations of effort ranging from K = 2/S = 150 to K = 20/S = 15 (R function based on Program SODA, Guillera-Arroita et al. 2010). We first considered the frequency of boundary estimates where \(\hat{\psi }\) = 1, and then dropped boundary estimates to calculate the A-optimality criterion. In a last step, we used our results from the optimum study design to calculate the conditional probability of site occupancy (\(\hat{\psi }_{condl}\)) that an alien species was actually present but overlooked at a site without detections (MacKenzie et al. 2006:100):
as well as the cumulative probability of detection (p*) that an alien species is recorded at least once in K visits to an occupied site (MacKenzie et al. 2006:95):
Ideally, a study design should have a low probability of conditional site occupancy but a high probability of cumulative detection.
Results
Study plots
We compiled six environmental covariates for the 60 study plots selected by using manual or random methods (Fig. 1). The 6.25 ha study plots varied in the percent cover of housing (median = 35.5%, range = 2.8–95.6%), roads (6.0%, 1.6–17.2%), forest (48.2%, 0.7–99.8%), and open fields (6.6%, 0–48.0%), annual rainfall (103.4 cm, 81.8–131.0), and population density (46.5 residents, 0–226). Pairwise correlations showed that the percent cover of housing was positively correlated with roads (+ 0.91) and population density (+ 0.29) but negatively correlated with forest cover (-0.68), open fields (-0.34), and annual rainfall (− 0.38, Fig. 3). Pairwise correlations between roads and forest or fields were also negative (− 0.38 to − 0.53). Since the percent cover of housing was correlated strongly with the other explanatory variables, we opted to use housing as an index of the local conditions at our study plots.
Vascular plants
A total of 239 alien species of vascular plants were recorded in 95 surveys of the 60 study plots in our 3-year study, 33 in 2018 only, 49 in 2019 only, 39 in 2020 only, 49 in 2 years, and 69 in all years (Table S2). We found alien species from five risk categories: no known impact (NK, n = 26 species), low impact (LO, n = 97), potentially high impact (PH, n = 32), high impact (HI, n = 29), and severe impact (SE, n = 55). The mean percentage of sites with at least one detection of the alien species (naïve occupancy or ψp) increased across the 5 risk categories: NK = 2.0% (range 1.7–5.0%), LO = 4.0% (1.7–25.0%), PH = 9.6% (1.7–65.0%), HI = 10.7% (1.7–40.0%), and SE = 25.0% (1.7–69.7%). The six most common species of alien plants detected in our study included Canadian goldenrod (Solidago canadensis SE) on 58 of 60 plots (96.7%), and red elderberry (Sambucus racemosa SE), yellow rocketcress (Barbarea vulgaris SE), Canadian fleabane (Conyza canadensis SE), shiny cotoneaster (Cotoneaster lucidus SE), and white sweetclover (Meliotosus albus SE) on 43–46 plots (65.0–76.7%).
We did not discover any new species of alien plants but we did find populations of 101 species that had ≤ 150 previous records in Norway (Artskart2). Six of these alien species have been assessed as high risk and our detections provide evidence of continuing establishment in Norway: gorse (Ulex europaeus HI, 1 of 60 plots, 1.7%), garden arabis (Arabis caucasica HI, 1.7%), contoneaster (Cotoneaster multiflorus HI, 1.7%), bearberry honeysuckle (Lonicera involucrata HI, 3 of 60 plots 5.0%), purple chokeberry (Aronia × prunifolia, 6 of 60 plots, 10.0%), and Virginia creeper (Parthenocissus quinquefolia HI, 10 of 60 plots, 16.7%). Other alien species with limited distributions have been assessed as low risk: pot marigold (Calendula officinalis NK, 1.7%), borage (Borago officinalis LO, 5.0%), and Himalayan blackberry (Rubus armeniacus LO, 5.0%). Last, we successfully detected a number of alien species of vascular plants with < 10 previous records in Norway. Examples of ‘doorknocker’ species that were each detected on one plot in our surveys included: highbrush blueberry (Vaccinium corymbosum LO), dwarf elder (Sambucus ebulus LO), max chrysanthemums (Leucanthemum maximum LO), hostas (Hosta ventricosa LO and H. lancifolia LO), and horned pansy (Viola × williamsii LO).
Invertebrates
We prepared samples for DNA-metabarcoding by crushing the insect biomass in 2018, and then by taking DNA extractions from the ethanol preservative in 2019. We conducted an initial validation of the ethanol method by taking a subset of 10 samples from 2019 and by sorting out all specimens from two Orders of insects: Coleoptera (ca. 4500 individuals) and Lepidoptera (ca. 1500 individuals). In both groups, ca. 20% of the specimens could not be identified by morphology because they were fragmentary or missing diagnostic structures. Of specimens that could be identified, roughly two-thirds of the coleopterans were false click beetles in the genus Trixagus which were not identified to species because of the large number of specimens. The remaining specimens were identified to species by morphology, which included 157 species of beetles and 210 species of butterflies or moths. Compared to the baseline diversity estimated by morphology, DNA extractions from crushed legs recovered a majority of taxa (Coleoptera: 76%, Lepidoptera: 92%). In contrast, DNA extractions from ethanol detected less than half of the species known to be present in the samples (Coleoptera: 35%, Lepidoptera: 46%). Accordingly, we conducted a second round of DNA extractions from animal tissue lysis (ATL) buffer for the remaining 70 samples from 2019. We had detected 1738 taxa from extractions from the filtered ethanol which increased by 30% to 2258 taxa for the extractions from lysis buffer, including an almost three-fold increase for Hymenoptera taxa (130–517 taxa). We combined information from all three methods of sample preparation in our subsequent analyses.
DNA metabarcoding generated 51,918,598 sequences, 47% thereof passed quality control, of which 64% were identified as arthropods (Table S3). PCR and DNA extraction negatives yielded predominantly non-insect sequences. Insect taxa detected in negative samples were removed from further analyses. We detected over 3500 taxa of arthropods from DNA-metabarcoding of 255 rounds of samples collected from 60 Malaise traps in our 3-year study. Of the total diversity, 135 species of arthropods were alien species of potential risk in Norway (Table S4). Twenty alien species were detected in four risk categories: low impact (LO, n = 12), potentially high impact (PH, n = 4), high impact (HI, n = 2), and severe impact (SE, n = 2). We detected no insects in the no known impact category (NK). In addition, we detected a large group of new species that had not been previously reported in Norway (NW, n = 115). The median percentage of sites with at least one detection of the alien species (ψp) covaried with risk category: NW = 3.3% (range 1.7–48.3%), LO = 3.3% (1.7–13.3%), and PH to SE = 9.2% (1.7–45.0%). The most common species of alien arthropods detected on our 60 plots included a harvestman (Opilio canestrinii SE, 48.3% of plots), the buff-tailed bumblebee (Bombus terrestris SE, 28.3%), a scavenger beetle (Cartodere nodifer PH, 20.0%), a plant bug (Deraeocoris lutescens HI, 13.3%), and a leaf weevil (Polydrusus formosus HI, 3.3%). We detected at least five alien species that have only recently arrived in Norway: a scavenger beetle (Cartodere bifasciata LO, 1.7%, first detected 2006), a gorse shield bug (Piezodorus lituratus HI, 1.7%, first detected 2006), a cicada (Tremulicerus fulgidus LO, 13.3%, first detected 2007), a leaf-mining moth (Cameraria ohridella LO, 5.0%, first detected 2013), and a false click beetle (Trixagus atticus LO, 6.7%, first detected 2016). Two species of alien insects with risk assessments that had not been previously found in the wild in Norway included a humpbacked fly (Dohrniphora cornuta LO, 3.3%) and a gall midge (Feltiella acarisuga LO, 1.7%). The 115 species of newly detected insects (NW) included 64 species of Diptera, including four species of flies in the genus Chalarus (NW, 3.3–48.3%), 25 species of Hymenoptera, including several species of parasitic wasps (Binodoxys brevicornis NW, 21.7%, Lysibia nanus NW, 16.7%, Aphidius uzbekistanicus NW, 15.0%), 15 species of Hemiptera, including three species of aphids (Myzus lythri NW, 5.7%; Aphis spiraecola NW, 2.9%; Cinara cupressi NW, 2.9%) and a leafhopper (Euscelidius variegatus NW, 1.7%) that are agricultural pests, and 4 species of Coleoptera, including an Asian beetle (Philonthus spinipes NW, 2.9%).
Occupancy analyses
The five top-ranked models in our candidate set for alien plants included the effects of risk category and year on the probability of detection, and the effects of risk category and housing on the probability of occupancy (Table 1). An effect of year was expected because we changed our survey methodology for alien plants from systematic transects in 2018 to a random search of study plots in 2019–2020. Estimates of the probability of detection for the second method tended to be higher in 2019–2020 for each risk category: NK/LO (2018: 0.137 ± 0.033; 2019: 0.188 ± 0.043; 2020: 0.159 ± 0.037), PH/HI (2018: 0.254 ± 0.034; 2019: 0.523± 0.048; 2020: 0.474 ± 0.047), and SE (2018: 0.602 ± 0.032; 2019: 0.702 ± 0.028; 2020: 0.650 ± 0.031, parameter estimates from model ψ(risk), p(risk×year). The probability of detection also covaried positively across risk categories and was higher for species of greater risk categories: NK/LO = 0.165, PH/HI = 0.427, and SE = 0.651 (Table 2). The probability of occupancy showed a similar pattern and was highest for species in the severe impact category: NK/LO = 0.144, PH/HI = 0.177, and SE = 0.318. Of the two most parsimonious models (ΔAICc ≤ 2.1), a model with an effect of housing on occupancy had 2.9× more support than a model without an effect (0.716/0.247). Parameter estimates from the minimum-AICc model showed that percent housing had a weak positive effect on the probability of occupancy, which increased slightly for the NK/LO and SE categories, but not for plants in the PH/HI category (Fig. 4A). Models with choice of site by random or manual criteria or observer effects received no support (wi < 0.001).
The four top-ranked models in our candidate set for alien insects included the effects of risk category on the probability of detection, and the effects of risk category and housing on the probability of occupancy (Table 1). Despite differences between years in the methods used in preparation of the insect samples for DNA extractions, models with year effects in the probability of detection received no support (wi < 0.001). The probability of detection was relatively low for three risk categories: NW = 0.207, LO = 0.326, and PH/HI = 0.127, and highest for the severe impact category: SE = 0.424 (Table 2). The probability of occupancy also covaried positively with risk category: NW = 0.115, LO = 0.052, PH/HI = 0.188, and SE = 0.422. Of the two parsimonious models (ΔAICc ≤ 2), a model with an effect of housing on occupancy had about half the support (0.49×) of a model without an effect (0.331/0.669). Similar to the results for alien plants, the relationship between probability of occupancy and percent housing was weakly positive for alien insects in the severe risk category, with higher rates of occupancy at study plots with more housing (Fig. 4B). Percent housing had less effect on occupancy of alien insects in the other risk categories. Again, models with choice of site by random or manual criteria or observer effects received no support (wi < 0.001).
Study design
We considered the optimum allocation of effort to number of visits per site (K) versus number of sites (S) for a national surveillance program based on a total sampling effort of TS ≈ 300 site-visits. Our three scenarios were based on mean parameter estimates for alien plants and insects at three levels of risk categories: a low risk scenario (LO: \(\overline{\psi }\) = 0.131 and \(\overline{p}\) = 0.206, based on estimates for NW, NK, and LO that were < 0.45), a high risk scenario (HI: \(\overline{\psi }\) = 0.250 and \(\overline{p}\) = 0.253, based on PH and HI), and severe risk scenario (SE: \(\overline{\psi }\) = 0.370 and \(\overline{p}\) = 0.301, based on SE, Fig. 5). The frequency of boundary estimates was > 10% for K ≤ 5 visits for the LO scenario, but was < 10% for all sampling designs for the HI and SE scenarios (Fig. 6A). To minimize variance for occupancy alone, 2–4 surveys per site would be sufficient for species of severe risk (SE) whereas 4–15 visits would be required for species in the lower risk categories (Fig. 5). The A-optimality criterion was calculated as the sum of variances for both occupancy and detection. The criterion was minimized with K = 15 visits to S = 20 sites for the LO scenario, K = 10 visits to S = 30 sites for the HI scenario, and K = 8 visits to S = 38 sites for the SE scenario (Fig. 6B). If we combined our mean estimates of occupancy and detection with the optimum number of visits, the conditional site occupancy (\(\hat{\psi }_{cond}\)) that an alien species was present at site without detections was consistently low (range = 0.005 to 0.032), whereas the cumulative probability of detection for an alien species (\(\hat{p}*\)) was high (range = 0.943–0.969).
Optimum number of replicate visits to be conducted at each sampling site where the criterion for study design is to minimize variance for estimates of occupancy (\(\hat{\psi }\)). Number of visits per site were calculated for a single-season occupancy model with equal costs per survey (based on Table 6.1 of MacKenzie et al. 2006). We generated three alternative scenarios based on our parameter estimates for alien plants and insects in Norway (Table 2)
Optimum number of replicate surveys to be conducted at each sampling site where the criteria for study design were: a to avoid boundary estimates, and b to minimize the combined variance for estimates of occupancy (\(\hat{\psi }\)) and detection (\(\hat{p}\), A-optimality). The three alternative scenarios were based on mean parameter estimates for alien plants and insects (LO: \(\overline{\psi }\) = 0.131, \(\overline{p}\) = 0.206; HI: \(\overline{\psi }\) = 0.250, \(\overline{p}\) = 0.253; SE: \(\overline{\psi }\) = 0.370, \(\overline{p}\) = 0.301) and a total sampling effort of TS ≈ 300 site-visits. Optimal sampling designs are marked with vertical arrows
Discussion
Our field project resulted in four major results. First, our systematic sampling design based on repeated sampling with a double-observer method (vascular plants) or with multiple rounds of sampling from Malaise traps (insects) were successful at detecting a large number of alien organisms. Even though many alien species were both rare and inconspicuous, we successfully recorded species that were new arrivals to Norway, species at an early stage of establishment, and cryptic species that may have been overlooked in previous surveys. Second, we demonstrated that DNA-metabarcoding is a promising technique for rapid processing and identification of invertebrates but that sample preparation based on crushing of insects or use of animal tissue lysis buffer recovered the highest percentage of taxa. Third, we found that the probabilities of occupancy and detection for alien species covaried with risk categories, with higher probabilities for organisms in the severe impact category. Last, we showed that our preliminary estimates can be used to design a study that optimizes the tradeoff between number of visits per site and number of sites to achieve a high confidence that an alien species would be detected with a systematic sampling program. We discuss the implications of our results for management of alien species and for development of a national surveillance program in Norway.
Discovery and origins of new alien species
Of the alien species detected in our sampling, none of the 239 species of vascular plants were new to Norway whereas 115 of 135 (85.2%) species of arthropods had not been previously detected in the country. Differences in new reports for the two groups of organisms likely reflect variation in sampling intensity, available taxonomic expertise, and challenges for identification of unfamiliar species (Sandvik et al. 2019). Many of the plant species detected in our sampling were thought to have a limited number of populations or had been recorded only a few times in Norway (Artskart2, Artsdatabanken 2020). At least six ‘doorknocker’ species were previously known from < 10 records, and our reports represent newly discovered populations. Highbrush blueberry (Vaccinium corybosum) is a cultivated plant that had not been previously reported from a natural site in Norway. Max chrysanthemums (Leucanthemum maximum), hostas (Hosta spp.), and horned pansy (Viola × williamsii) are all introduced garden plants that have been occasionally reported from sites with dumping of garden refuse. Dwarf elder (Sambucus ebulus) is another garden plant that was first found in 2007 near Vindafjord municipality in Rogaland county. Our unexpected discovery of a relatively large stand of plants (64 m2) nearly 400 km away at Hurumlandet in Viken county could be a long-distance dispersal event or may indicate that the species is more widely established than previously known.
The uncommon alien plants that were detected in our project also varied in their origins and ecology (Artsdatabanken 2020). Gorse (Ulex europaeus) is a thorny shrub that was first introduced to Norway in transport of soil and timber but has been present for more than a century with limited spread. Virginia creeper (Parthenocissus quinquefolia) is an alien species that has been distributed for planting by the Norwegian Public Roads Administration. Several species are cultivated as vegetables (Borago officinalis) or flowering plants (Arabis causcasica, Calendula officinalis), but have escaped from gardens and in some cases are now expanding in natural areas. Hostas (Hosta ventricosa) were first discovered at dumping site for garden refuse. Purple chokeberry (Aronia x prunifolia), bearberry honeysuckle (Lonicera involucrata), and Himalayan blackberry (Rubus armeniacus) are perennial shrubs with edible berries that have been planted as garden ornamentals or for fruit production, and expansion into natural areas could be due to seed dispersal by fruit-eating birds (Bennett et al. 2011). Himalayan blackberries are perhaps the species of greatest concern since they were first discovered in Norway in 1953, spread to another 50 sites by 1989, and our new records from three study plots are evidence of continuing range expansion.
Our detections of alien insects included two species not previously found in the wild in Norway, a humpbacked fly (Dohrniphora cornuta) and a gall midge (Feltiella acarisuga). D. cornuta was first collected from a cadaver in Sweden by forensic entomologists in 2012 (Disney et al. 2014), and our detections on two plots are the first records for Norway. The gall midge F. acarisuga has been used in Norway for biological control of spider mites (Tetranychus spp.) in greenhouses (Artsdatabanken 2020), but our detections are the first record of the species in the wild. We also detected a suite of alien insects from continental Europe that are relatively new arrivals in Norway. The cicada Tremulicerus fulgidus requires black poplar (Populus nigra) as a host plant and was first found in Sweden in 2005 and then in Norway in 2007 (Endrestøl 2008). Similarly, the leaf-mining moth Cameraria ohridella feeds upon leaves of the European horse-chestnut (Aesculus hippocastanum) and was first detected in Norway in 2013 (Aarvik et al. 2014). The false click beetle Trixagus atticus has only been previously found south of our study area and was first detected at a waste disposal site near Frederikstad in 2016 (Artsdatabanken 2020).
We detected 115 new species of alien insects and other arthropods that had not been previously reported in Norway. A majority of the new species belong to taxonomic groups that have not been well-studied in Norway, including small-bodied species in the orders Diptera (n = 70 species), Hymenoptera (n = 26) and Hemiptera (n = 18). Some of these taxa were from poorly known groups that are particularly difficult to identify from morphology, such as the parasitic flies in the genus Chalarus (Kehlmeier and Assman 2008). Of the 15 new species detected on more than 20% of our study plots, at least 3 species had been previously reported from Sweden (Contacyphon ruficeps, Procladius ferrugineus, Binodoxys brevicornis), suggesting that some might be native species that have been previously overlooked (Karlsson et al. 2020). Nevertheless, several of the new species are also known to be alien species in Europe, including the Asian rove beetle Philonthus spinipes (Schillhammer 1999). At least two newly detected aphids are economic pests: Aphis spiraecola is a pest of apples and citrus fruits, whereas Cinara cupressi is a pest of cypress and other conifers (Watson et al. 1999; Tsai and Wang 2001). A third aphid species Myzus lythri occurs Sweden where it uses two alien plants as alternate hosts: loosestrife (Lythrum virgatum NK) and mahaleb cherry (Prunus mahaleb NK, Heie 2009). Both species of alien plants were detected in our project, but at different study plots than our records for M. lythri.
Challenges with DNA-metabarcoding for insects
We tested four alternative methods for identification of insects collected in our surveillance program. DNA-metabarcoding was cost-effective and more efficient than sorting and processing samples by hand. However, comparisons of methodology showed that the DNA metabarcoding method missed some species of Coleoptera and Lepidoptera that were detected with traditional methods based on morphological identification (Jacobsen et al. 2020). Underestimates of diversity may have been due to problems of insufficient DNA, marker failure, and poor database coverage (Comtet et al. 2015). We expected that detection rates might vary among species relative to variation in body size, surface area to volume ratio, and degree of sclerotization (Marquina et al. 2019). The COI barcode region usually showed sufficient divergence to distinguish between closely related species, but also failed in some cases. Here, DNA-metabarcoding gave preliminary detection of an alien marsh beetle Cyphon ruficeps, but morphological identification from specimens determined that the species was actually a native congener C. coarctatus. Further investigation of reference sequence data showed that the COI barcode cannot distinguish between the congeneric species of Cyphon. Last, we found that for some poorly studied but species-rich lineages (Diptera, Hymenoptera), several thousand species reported from Norway still lack publicly available reference sequences for the COI barcode region, and thus cannot yet be identified to species with DNA-metabarcoding.
Of the three metabarcoding approaches we tested, DNA extractions from the ethanol preservative gave the worst results, recovering only 35–46% of the beetle and butterfly species identified with morphological criteria. Previous studies have used DNA-extractions from ethanol to recover 65–100% of aquatic communities of mayflies, stoneflies, and caddisflies (Hajibabaei et al. 2012; Carew et al. 2018; Zizka et al. 2019). In our study, ethanol extractions could have been less successful because exoskeletons of terrestrial insects have greater sclerotization and a thicker cuticle that may have reduced leaching of DNA from the body tissues. Aquatic samples might also be more likely to include soft-bodied larval or nymphal life-stages, whereas Malaise traps are more likely to sample flying adults in terrestrial habitats. DNA extractions from crushed animal parts and from animal tissue lysis (ATL) buffer both led to a dramatic increase in the number of taxa recovered, especially for Hymenoptera. We recommend the use of extractions from lysis buffer because it effectively recovers a majority of taxa and is also a non-destructive method where specimens were retained for later validation. Access to physical specimens allows for confirmation of new finds and supplemental identification using morphological characters, which avoids some of the analytical problems associated with the genetic methods. For example, we first detected the invasive rove beetle Philonthus spinipes using DNA-metabarcoding of a lysis sample and then successfully confirmed the new record from inspection of a preserved specimen retained in our insect samples.
Occupancy models and sampling designs for alien species
Occupancy models are becoming a valuable tool for determining the probability that alien species are present in a sampling plot while controlling for imperfect detection (Tavecchia et al. 2017; Davis et al. 2018; Fossøy et al. 2020). The different axes of ψ–p parameter space can be used to evaluate whether alien species are rare or common (low to high occupancy), and cryptic or conspicuous (low to high detection, Guillera-Arroita et al. 2010; Reich 2020). Our occupancy analyses for alien species in Norway showed that the ranked risk assessment categories had different combinations of probabilities: new alien species were rare and cryptic, severe impact species were more common and conspicuous, whereas high impact species had intermediate rates. Risk assessments for alien species in Norway are based on multiple ecological criteria, including invasion potential, impacts on native or threatened species, and impacts on sensitive ecosystems (Sandvik et al. 2020). However, two of the criteria for assessing invasion potential include the rate of range expansion and the percentage of the landscape that is occupied. Thus, the alien species categorized as severe impact likely had high occupancy and detection probabilities because they are already well-established and recognizable species. If a project goal is to minimize variance of occupancy alone, then our current sampling design with 2–4 rounds of sampling would be adequate for plants and insects of severe risk. On the other hand, if the project goal is to minimize variance for both occupancy and detection rates, then the optimal sampling design for our severe risk scenario would require more intensive sampling with K = 8 visits to S = 38 sites if TS = 300.
A national surveillance program might be best targeted at detection of new alien species where eradication is more feasible at the early stages of establishment. Unfortunately, our results show that new alien species will be particularly challenging to discover because they are in a ψ–p parameter space where they are both rare and cryptic. The probabilities of occupancy and detection were low in our low risk scenario, and sampling designs with few visits to many sites would lead to a high frequency of boundary estimates where ψ = 1. The optimal sampling designs for our low to high risk scenarios had K = 10–15 visits to S = 20–30 sites which would be a different allocation of effort than our current sampling scheme. Linkages to ecological covariates can help with study design for selecting sites where novel alien species are more likely to occur. We tested site selection based on the hotspot model of Olsen et al. (2017) versus known sites with transport hubs or recycling depots but found that occurrence and detection of alien species was similar between random and manually selected sites. Percent housing was a good index of local conditions at our study plots, and the weak positive relationships between occupancy and percent housing for alien plants and insects were consistent with domestic gardens, garden refuse and other types of anthropogenic disturbance as a source of introductions (Fremstad and Elven 1997; Westergaard et al. 2018).
Different classes of occupancy models could provide additional insights in surveillance programs for alien organisms. Here, we conducted surveys for multiple species of invasive organisms and therefore used a ‘standard’ occupancy design with S sites each visited K times. Reich (2020) showed that the standard design has the best performance if ψ = 0.2–0.3 and p = 0.1–0.7, which was the case in our study. Alternative occupancy designs include ‘removal’ models where S sites are visited K times but repeated visits are discontinued after a positive detection, and ‘conditional’ designs where S sites are visited once and sites with positive detections are then visited an additional K−1 times. The two alternative designs are better suited for surveillance of single alien species but can give better performance if the species is either common (ψ ≥ 0.4, removal) or rare (ψ ≤ 0.2, conditional). In a national surveillance program aimed at early detection of alien species, false positives due to species misidentification should be a lower concern than false absences. Here, we used single-season occupancy models to control for false negatives and minimized false positives with a double-observer method for plants and by confirming species identity from insect specimens. False positives can also be controlled with multistate occupancy models with different codes for certainty of detection (Miller et al. 2011; Ficetola et al. 2015). Last, we used occupancy models based on fixed effects so that candidate models could be fit quickly with maximum likelihood methods and ranked by AICc. Multi-species occupancy models in a Bayesian framework would have the added advantage of information sharing by treating species as a random effect (Iknayan et al. 2014).
Conclusion
We successfully completed the first step of developing an Early Detection and Rapid Response (EDRR) system for alien plants and insects in Norway. Our field surveys and DNA-metabarcoding were effective at providing early detections of alien organisms that were new arrivals or at an early stage of establishment. For study plots where our sampling revealed the first detections of doorknocker species, follow-up surveys will need to be used to check for potential spread. To be more efficient at detection of alien species that are rare and cryptic, better methods for site selection need to be developed and sampling effort might be reallocated to more visits per site. Sample preparation methods had a large effect on the success of DNA-metabarcoding and we recommend DNA extractions based on lysis buffer for terrestrial arthropods. Our bioinformatic pipeline provided a fast and efficient method for taxonomic assignment but existing databases remain incomplete for Norway and expanding content for both native and alien species should be a future priority. Jacobsen et al. (2020) showed that the financial costs of surveillance efforts would likely be offset by reducing the potential costs of damage from alien species to ecosystem services, human health, and infrastructure. In the future, development of tools to implement a rapid response will require public engagement, coordination of regional authorities, and prioritization of resources for eradication of alien organisms (Qvenild et al. 2014; Reaser et al. 2020).
Data availability
Occurrence records for alien plants and insects have been published at GBIF Norway (https://doi.org/10.15468/q3abt7) and on the Species Map of the Norwegian Biodiversity Information Centre (Artskart2, artskart.artsdatabanken.no). Barcodes for newly discovered species of insects in Norway have been submitted to the Barcode of Life Data database (BOLD) via the portal for the Norwegian Barcode of Life (NorBol, norbol.org). The Eukaryote CO1 reference set for the RDP classifier is archived at Zenodo (https://doi.org/10.5281/zenodo.4741447). Derived datasets and R scripts for the statistical analyses are available at the Dryad digital repository (https://doi.org/10.5061/dryad.0rxwdbs4f).
Change history
24 January 2023
Missing Open Access funding information has been added in the Funding Note
References
Aarvik L, Boumans L, Sørlibråten O (2014) The horse chestnut leaf-miner, Cameraria ohridella Deschka & Dimić, 1986, (Lepidoptera, Gracillariidae) established in Norway. Nor J Entomol 61:8–10
Alpert P (2006) The advantages and disadvantages of being introduced. Biol Invasions 8:1523–1534
Artsdatabanken (2015) Artsnavnebase 2015: Species nomenclature database (In Norwegian). The Norwegian Biodiversity Information Centre (Artsdatabanken) Trondheim, Norway
Artsdatabanken (2020) Fremmedartslista 2018: Alien species in Norway (In Norwegian). The Norwegian Biodiversity Information Centre (Artsdatabanken) Trondheim, Norway
Åström J, Birkemoe T, Dahle S et al (2022) Insect monitoring in Østlandet and Trøndelag. Report from field season 2021 (In Norwegian). NINA report 2070. Norwegian Institute for Nature Research, Trondheim, Norway
Bebber DP, Ramotowski MAT, Gurr SJ (2013) Crop pests and pathogens move polewards in a warming world. Nat Clim Chang 3:985–988
Bellard C, Jeschke JM, Leroy B et al (2018) Insights from modeling studies on how climate change affects invasive alien species geography. Ecol Evol 8:5688–5700
Bennett JR, Young EJ, Giblin DE et al (2011) Avian dispersal of exotic shrubs in an archipelago. Ecoscience 18:369–374
Blaalid R, Often A (2019) Alien species list 2018: with focus on vascular plants (In Norwegian). Naturen 6:250–256
Bohmann K, Elbrecht V, Caroe C et al (2022) Strategies for sample labelling and library preparation in DNA metabarcoding studies. Mol Ecol Resour 22:1231–1246
Braukmann TWA, Ivanova NV, Prosser SWJ et al (2019) Metabarcoding a diverse arthropod mock community. Mol Ecol Resour 19:711–727
Bryn A, Strand G-H, Angeloff M et al (2018) Land cover in Norway based on an area frame survey of vegetation types. Nor J Geogr 72:131–145
Callahan BJ, McMurdie PJ, Rosen MJ et al (2016) DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583
Carew ME, Coleman RA, Hoffmann AA (2018) Can non-destructive DNA extraction of bulk invertebrate samples be used for metabarcoding? PeerJ 6:e4980
Chapman D, Purse BV, Roy HE et al (2017) Global trade networks determine the distribution of invasive non-native species. Glob Ecol Biogeogr 26:907–917
Comtet T, Sandionigi A, Viard F et al (2015) DNA (meta)barcoding of biological invasions: a powerful tool to elucidate invasion processes and help managing aliens. Biol Invasions 17:905–922
Darling JA, Blum MJ (2007) DNA-based methods for monitoring invasive species: a review and prospectus. Biol Invasions 9:751–765
Davis AJ, McCreary R, Psiropoulos J et al (2018) Quantifying site-level usage and certainty of absence for an invasive species through occupancy analysis of camera-trap data. Biol Invasions 20:877–890
Disney RH, Garcia-Rojo A, Lindstrom A et al (2014) Further occurrences of Dohrniphora cornuta (Bigot) (Diptera, Phoridae) in forensic cases indicate likely importance of this species in future cases. Forensic Sci Int 241:e20-22
Elbrecht V, Braukmann TWA, Ivanova NV et al (2019) Validation of COI metabarcoding primers for terrestrial arthropods. PeerJ 7:e7745
Elbrecht V, Peinert B, Leese F (2017) Sorting things out: Assessing effects of unequal specimen biomass on DNA metabarcoding. Ecol Evol 7:6918–6926
Endrestøl A (2008) Hoppers on black poplars–The Auchenorrhyncha fauna on Populus nigra in Norway. Nor J Entomol 55:137–148
Endrestøl A, Økland B (2019) Alien insects in forest and garden (In Norwegian). Naturen 6:257–271
Ficetola GF, Pansu J, Bonin A et al (2015) Replication levels, false presences and the estimation of the presence/absence from eDNA metabarcoding data. Mol Ecol Resour 15:543–556
Fremstad E, Elven R (1997) Alien plants in Norway and dynamics in the flora: a review. Nor J Geogr 51:199–218
Fossøy F, Brandsegg H, Sivertsgård R et al (2020) Monitoring presence and abundance of two gyrodactylid ectoparasites and their salmonid hosts using environmental DNA. Environ DNA 2:53–62
Guillera-Arroita G, Ridout MS, Morgan BJT (2010) Design of occupancy studies with imperfect detection. Methods Ecol Evol 1:131–139
Hajibabaei M, Spall JL, Shokralla S et al (2012) Assessing biodiversity of a freshwater benthic macroinvertebrate community through non-destructive environmental barcoding of DNA from preservative ethanol. BMC Ecol 12:art28
Heie OE (2009) Aphid mysteries not yet solved (Hemiptera: Aphidomorpha). Aphids Hemipterous Insects 15:49–60
Hulme PE (2009) Trade, transport and trouble: managing invasive species pathways in an era of globalization. J Appl Ecol 46:10–18
Iknayan KJ, Tingley MW, Furnas BJ et al (2014) Detecting diversity: emerging methods to estimate species diversity. Trends Ecol Evol 29:97–106
Jacobsen RM, Endrestøl A, Magnussen K, et al (2020) Early detection and warning of new alien species in Norway: Testing and developing surveillance of alien terrestrial vascular plants and insects (In Norwegian). NINA Report 1729. Norwegian Institute for Nature Research, Trondheim, Norway
Karlsson D, Hartop E, Forshage M et al (2020) The Swedish Malaise Trap project: a 15 year retrospective on a countrywide insect inventory. Biodivers Data J 8:e47255
Kehlmaier C, Assmann T (2008) The European species of Chalarus Walker, 1834 revisited (Diptera: Pipunculidae). Zootaxa 1936:1–39
Kopylova E, Navas-Molina JA, Mercier C et al (2016) Open-source sequence clustering methods improve the state of the art. mSystems. https://doi.org/10.1128/mSystems.00003-15
Lodge DM, Williams S, MacIsaac HJ et al (2006) Biological invasions: recommendations for U.S. policy and management. Ecol Appl 16:2035–2054
Lyal CHC, Miller SE (2020) Capacity of United States federal government and its partners to rapidly and accurately report the identity (taxonomy) of non-native organisms intercepted in early detection programs. Biol Invasions 22:101–127
MacKenzie D, Nichols J, Royle J et al (2006) Occupancy estimation and modeling: inferring patterns and dynamics of species occurrence. Academic Press, Burlington
Marquina D, Esparza-Salas R, Roslin T et al (2019) Establishing arthropod community composition using metabarcoding: surprising inconsistencies between soil samples and preservative ethanol and homogenate from Malaise trap catches. Mol Ecol Resour 19:1516–1530
Martin M (2011) Cutadapt removes adapter sequences form high-throughput sequencing reads. EMBnet J 17:10–12
Meyerson LA, Mooney HA (2007) Invasive alien species in an era of globalization. Front Ecol Environ 5:199–208
Milbau A, Stout JC (2008) Factors associated with alien plants transitioning from casual, to naturalized, to invasive. Conserv Biol 22:308–317
Miller DA, Nichols JD, McClintock BT et al (2011) Improving occupancy estimation when two types of observational error occur: non-detection and species misidentification. Ecology 92:1422–1428
NOBANIS (2019) European network on invasive alien species. www.nobanis.org, Accessed on Dec 1, 2019
Olsen SL, Åström J, Hendrichsen D, et al. (2017) Alien vascular plants in Norway: modelling of introduction areas and current distribution (In Norwegian). NINA Report 1393. Norwegian Institute for Nature Research, Oslo, Norway
Otto CRV, Bailey LL, Roloff GJ (2013) Improving species occupancy estimation when sampling violates the closure assumption. Ecography 36:1299–1309
Porter TM (2017) Eukaryote CO1 reference set for the RDP classifier (Version v. 4.0.1). Zenodo. https://doi.org/10.5281/zenodo.4741447
Porter TM, Hajibabaei M (2018) Automated high throughput animal CO1 metabarcode classification. Sci Rep 8:4226
Qvenild M, Setten G, Skår M (2014) Politicising plants: dwelling and invasive alien species in domestic gardens in Norway. Nor J Geogr 68:22–33
R-Core-Team (2020) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria (URL: www.R-project.org)
Reaser JK, Burgiel SW, Kirkey J et al (2020) The early detection of and rapid response (EDRR) to invasive species: a conceptual framework and federal capacities assessment. Biol Invasions 22:1–19
Reich HT (2020) Optimal sampling design and the accuracy of occupancy models. Biometrics 76:1017–1027
Reichard SH, White P (2001) Horticulture as a pathway of invasive plant introductions in the United States. Bioscience 51:103–113
Rota CT, Fletcher RJ Jr, Dorazio RM et al (2009) Occupancy estimation and the closure assumption. J Appl Ecol 46:1173–1181
Sakai AK, Allendorf FW, Holt JS et al (2001) The population biology of invasive species. Annu Rev Ecol Syst 32:305–332
Sandvik H, Dolmen D, Elven R et al (2019) Alien plants, animals, fungi and algae in Norway: an inventory of neobiota. Biol Invasions 21:2997–3012
Sandvik H, Hilmo O, Henriksen S et al (2020) Alien species in Norway: results from quantitative ecological impact assessments. Ecol Solut Evid 1:e12006
Schillhammer H (1999) Revision of the East Palaearctic and Oriental species of Philonthus Stephens-Part 2. The spinipes and cinctulus groups (Coleoptera: Staphylinidae, Staphylininae). Koleopterologische Rundschau 69:55–65
Simberloff D (2003) How much information on population biology is needed to manage introduced species? Conserv Biol 17:83–92
Sutherland S (2004) What makes a weed a weed: life history traits of native and exotic plants in the USA. Oecologia 141:24–39
Tavecchia G, Miranda MA, Borras D et al (2017) Modelling the range expansion of the Tiger mosquito in a Mediterranean Island accounting for imperfect detection. Front Zool. 14:art39 https://doi.org/10.1186/s12983-017-0217-x
Tsai JH, Wang J-J (2001) Effects of host plants on biology and life table parameters of Aphis spiraecola (Homoptera: Aphididae). Environ Entomol 30:44–50
Wang Q, Garrity GM, Tiedje JM et al (2007) Naïve Bayesian classifier for rapid assignment of rRNA Sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267
Wang WY, Srivathsan A, Foo M et al (2018) Sorting specimen-rich invertebrate samples with cost-effective NGS barcodes: validating a reverse workflow for specimen processing. Mol Ecol Resour 18:490–501
Wasof S, Lenoir J, Aarrestad PA et al (2015) Disjunct populations of European vascular plant species keep the same climatic niches. Glob Ecol Biogeogr 24:1401–1412
Watson GW, Voegtlin DJ, Murphy ST et al (1999) Biogeography of the Cinara cupressi complex (Hemiptera : Aphididae) on Cupressaceae, with description of a pest species introduced into Africa. Bull Entomol Res 89:271–283
Westergaard KB, Endrestøl A, Hanssen O, et al. (2018) Alien stowaway species: pathways of import in horticultural plants. Monitoring and method development 2017–2018 (In Norwegian). NINA Report 1557. Norwegian Institute for Nature Research, Trondheim, Norway
Yu DW, Ji Y, Emerson BC et al (2012) Biodiversity soup: metabarcoding of arthropods for rapid biodiversity assessment and biomonitoring. Methods Ecol Evol 3:613–623
Zizka VMA, Leese F, Peinert B et al (2019) DNA metabarcoding from sample fixative as a quick and voucher-preserving biodiversity assessment method. Genome 62:122–136
Ødegaard F, Tømmerås BÅ (2000) Compost heaps: refuges and stepping-stones for alien arthropod species in northern Europe. Divers Distrib 6:45–59
Acknowledgements
We thank H. Rui for assistance with registration of alien plants, and K. Berggren and O. Hanssen for help with identification of beetles and butterflies. T. Holmern provided cooperation and support as our main contact at the Norwegian Environmental Agency. Insect sampling at the Ekebergskråningen nature reserve was conducted under permits from the County Governor of Oslo and Akershus. We are grateful to the individuals and businesses who gave us permission to conduct field sampling on private and public lands, including Borregaard AS, Bane NOR, Drammen Harbor, Eventyrhus AS, Movar IKS, Ragn Sells AS, RfD IKS, Statkraft Tofte AS, Statskog Glomma AS, Sætre Bruk AS, and the municipalities of Drammen, Nittedal, and Oslo. Two anonymous referees provided constructive feedback on an earlier draft of the manuscript.
Funding
Open access funding provided by Norwegian institute for nature research. The field project was supported with research funding under three contracts from the Norwegian Environmental Agency (M-1142|2018, M-1576|2019, and M-1855|2020). The research was also partly financed by basic funding to the Norwegian Institute for Nature Research from the Research Council of Norway (Project No. 160022/F40). Funding for open access publication was provided under a national agreeement established by the Norwegian Agency for Shared Services in Education and Research.
Author information
Authors and Affiliations
Contributions
RMJ, AE, JÅ and RB were responsible for developing and coordinating the field project. AO and HH conducted the field surveys for alien plants. AE and RMJ conducted the field surveys for insects, and AE completed the morphological identification of beetles and butterflies. FF, MLD and MAMM conducted the DNA metabarcoding and developed a new bioinformatics pipeline for taxonomic identification. BKS and JÅ conducted the statistical analyses. BKS and MLD wrote the manuscript with contributions from the coauthors.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts or competing interests to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sandercock, B.K., Davey, M.L., Endrestøl, A. et al. Designing a surveillance program for early detection of alien plants and insects in Norway. Biol Invasions 25, 917–936 (2023). https://doi.org/10.1007/s10530-022-02957-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10530-022-02957-6