Abstract
Polyploidy may contribute to invasive ability as it can lead to high survival and fitness during establishment and enhance the processes of adaptation to novel environments by increasing genetic diversity in invading propagules. Many grasses are polyploid and many are aggressive invaders, making them persistent problems in disturbed environments worldwide. Today, vast areas of central Panama are dominated by Saccharum spontaneum, a perennial grass that originates from Asia. While widely regarded as invasive, it is not known when or how it arrived in Panama. We explore hypotheses regarding the timing and origins of this invasion through literature review and comparisons of genetic diversity in Panama with accessions from available sugarcane germplasm collections, highlighting historical accessions that were likely brought to Panama in 1939 as part of a USDA sugarcane germplasm collection. Samples were haplotyped at two chloroplast loci and genotyped using eight microsatellite markers. All sequenced individuals from Panama belong to a single chloroplast lineage which is common worldwide and was common in the Historic germplasm collection. Although genotypic diversity was extremely high in all samples due to high ploidy, samples from Panama had reduced diversity and clustered with several accessions in the Historic collection which had the same haplotype and high ploidy levels. Our results suggest that accidental escape from the historical sugarcane germplasm collection is the likely origin of the S. spontaneum invasion in Panama. Intraspecific hybridization among several historical accessions and pre-adaptation to local conditions may have facilitated its rapid spread and persistence. We discuss the implications of our findings for biosecurity of germplasm collections.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Grasses have been identified as a particularly invasive plant group and are overly represented in “invasive alien plant” lists globally (D’Antonio et al. 2011; Holm et al. 1977). Invasive grasses are a persistent problem in tropical ecosystems worldwide as many have been introduced for forage and as improved pasture grasses and have escaped agricultural settings, now occupying many disturbed habitats and posing a threat to native communities (D’Antonio and Vitousek 1992; Parsons 1972; Williams and Baruch 2000). Many are large-statured and form monocultures, excluding other species and, in many cases, rapidly transforming the landscapes that they invade (Lambert et al. 2014; Canavan et al. 2019). In the wet Neotropics, invasive grasses have increased fire regimes (Saltonstall and Bonnett 2012; Williams and Baruch 2000) and inhibited natural regeneration of forest communities across the region (Boeschoten et al. 2020; Hooper et al. 2005; Jones et al. 2004; Wishnie et al. 2002; Nepstad et al. 1991).
Genetic diversity is predicted to facilitate invasions by increasing potential for rapid evolution and adaptation to new environments (Dluglosch et al. 2015; Dlugosch and Parker 2008; Sakai et al. 2001). However, genetically depauperate invaders are also known (e.g. Japanese knotweed (Fallopia japonica [Hollingsworth and Bailey 2000], fountain grass (Pennisetum setaceum [Le Roux et al. 2007], giant reed (Arundo donax [Ahmad et al. 2008; Hardion et al. 2014]), and a multitude of traits such as broad environmental tolerance (Le Roux et al. 2007), phenotypic plasticity (Richards et al. 2006), escape from natural enemies (Parker and Gilbert 2007), presence of pre-adapted genotypes (Hurka et al. 2003), and ability to disperse and germinate rapidly (Gioria and Pyšek 2017) have been proposed to also contribute to the success of species when establishing in new environments. Although introductions often begin with only a few propagules, high ploidy levels can facilitate invasions by increasing genetic diversity in the invading propagules, decreasing the forces of founder effects and genetic drift, and making invaders more tolerant to selfing due to reduced inbreeding depression (te Beest et al. 2012; Pandit et al. 2011; Soltis and Soltis 2000). Further, intraspecific hybridization of introduced lineages from geographically distinct source populations can both increase genetic diversity and facilitate adaptation to new environments by creating novel combinations of alleles and potentially stimulating hybrid vigor (Dluglosch et al. 2015; Ellstrand and Schierenbeck 2000; Lavergne and Molofsky 2007; Novak and Mack 2005).
Saccharum spontaneum L. (wild sugarcane) is a perennial C4 grass that occupies a diversity of habitats and has a broad native range, extending from Africa (e.g. Kenya, Uganda, Egypt, Algiers) through temperate Asia (e.g. Israel, Saudi Arabia, Afghanistan, Pakistan, China, Japan) and tropical Asia (e.g. India, Indonesia, Malaysia, Papua New Guinea, Thailand, Vietnam) (Panje and Babu 1960). It is an erect, sometimes rhizomatous, grass that is highly polymorphic across its range, ranging in morphology from a small stalkless bunchgrass to 6 m tall stalks. When flowering, it forms an abundance of white inflorescences which produce large numbers of seeds that are wind dispersed. Ploidy levels vary within the species (2n = 40–128) and genetic diversity is high across ploidy levels (Aitken et al. 2018; Brown et al. 2007; Cordeiro et al. 2000) which tend to be higher in tropical regions than in the subtropics and temperate regions (Brandes and Sartoris 1936). Reproduction may be purely asexual or largely sexual, depending on its growth form (Panje 1970). It has been naturalized in many locations, including France, Panama, Costa Rica, Cuba, Australia, and the United States (USDA 2016; Westbrooks and Miller 1993) and has been reported as a weed in 33 countries (Holm et al. 1977).
In central Panama, S. spontaneum today occupies 2–3% of the land cover of the Panama Canal Watershed (PCW) (ANAM-ACP 2006) where it has invaded abandoned agricultural lands and is common along roadsides (Cerezo 2010; Hammond 1999). It is rhizomatous and forms dense grasslands that exclude other plants from establishing and its growth is stimulated by the dry-season fires that now occur annually in the PCW (Hammond 1999; Hooper et al. 2005; Saltonstall and Bonnett 2012). It produces large quantities of seed which have high germinability and have likely facilitated its rapid spread through the PCW and surrounding areas (Bonnett et al. 2014). Although a number of large-statured invasive grasses are found in the PCW today, S. spontaneum is the most aggressive (K. Saltonstall, pers. obs).
Although it is not known exactly when S. spontaneum was introduced to Panama, the first herbarium accessions date to the early 1960’s (TROPICOS, STRI/SCZ herbarium). However, botanical records note its presence and rapid spread as early as the mid-1940s (Moore 1948). There are a number of theories regarding how S. spontaneum was introduced to Panama, including its use for erosion control during construction of the Panama Canal in the early 1900 (von Lindeman 1986), transport of vegetative fragments via ships carrying construction equipment from Asia through the Panama Canal in the 1970’s (Hammond 1999), and accidental escape from the US Sugarcane Germplasm collection (C. Elton, personal communication). The first two theories, while compelling, are unlikely. Numerous photographs still exist from the time during and decades following the construction of the Panama Canal and while extensive grasslands are often seen along the banks of the Canal, those taken during the flowering season suggest that Panicum maximum Jacq. (guinea grass) was likely the dominant grass along the Canal in the first half of the 20th century (K. Saltonstall, personal observation). Similarly, while there were military bases at either ends of the Canal and heavy equipment was surely transported to Panama on ships, both observations and actual collections of S. spontaneum specimens pre-date the 1970’s. In addition, experiments testing the growth of rhizome fragments have shown small fragments to grow very poorly, even when fresh tissues are used (Bonnett et al. 2014).
In 1939, the U.S. Department of Agriculture (USDA) reference collection of sugarcane varieties was brought to the Canal Zone Experimental Gardens (CZEG), then located in what is now known as Summit Gardens in the PCW, in part to facilitate crossing of some sugarcane varieties that bloom at different times of the year at different latitudes and to protect the collection from possible hurricane damage (CZEG 1939). The collection included about 500 accessions of ancestral forms of sugarcane as well as varieties cultivated in areas where sugarcane is indigenous and other genera closely related to Saccharum (CZEG 1939). It is not known how long the collection remained in Panama but experimental studies suggest that it was there for at least five years (1939–1944; Brandes 1950), but likely not much longer (Croat 1971). It is also unknown how many of the accessions in the collection were S. spontaneum as these records were destroyed in a fire. However, in 1938 a list of 25 S. spontaneum accessions making up the USDA collection were described (Brandes et al. 1938), of which seven are known to have been used in experimental growth studies at the CZEG (Brandes 1950).
This study investigates whether the USDA sugarcane reference collection was the most likely source of the S. spontaneum invasion in Panama using a phylogeographic approach. We assess levels of genetic diversity in the modern population in Panama using chloroplast DNA (cpDNA) sequencing and microsatellite markers and compare allele frequencies with accessions from the modern sugarcane germplasm collection (World collection) that originate throughout the range of S. spontaneum. We then look at genetic diversity of a subset of accessions that were likely present in the sugarcane reference collection in 1939 (Historic collection) based on the list described by Brandes (1939) and USDA import records (USDA-GRIN 2009) and compare these with the modern Panamanian population. We discuss the implications of our findings with regard to pre-adaptation of potential invaders to novel environments and biosecurity of germplasm collections.
Materials and methods
Study location
The Republic of Panama (9° N, 79° W) is in the moist tropics of central America, bordered by the Caribbean Sea to the north and the Pacific Ocean to the south. Rainfall is seasonal, with a 7–8 month rainy season and a dry season from December to April. The Panama Canal watershed (PCW) includes about 2892 km2 of land area in central Panama (Condit et al. 2001) and most of the human population is concentrated at lower elevations. On the drier Pacific side much of the forest cover was cleared during construction of the Panama Canal and, beginning in the 1950’s, for cattle pastures. Widespread abandonment of agricultural lands in the 1970’s led to S. spontaneum invasion into many of these open areas (Condit et al. 2001; Heckadon-Moreno et al. 1999).
Sample collection
Leaf samples were collected throughout Central Panama and nearby locations (n = 196 samples) from 2009 to 2010 (see Supplementary Table S1 in Supporting Information). Saccharum spontaneum occurs both in discrete patches and in large continuous stands along roadways, stream channels, and abandoned agricultural lands throughout the region. Single leaf samples were collected from the majority of stands but samples were also collected along transects at 10 m intervals at 22 sites to assess the extent of clonal diversity within these stands (Bonnett et al. 2014). All samples were kept frozen at − 20 °C until processed in the laboratory where DNA was extracted using a modified CTAB extraction protocol (Doyle and Doyle 1987).
DNA from the world germplasm collection
Leaf tissue was obtained from the world sugarcane germplasm collection as in Aitken et al. (2018). All accessions used are listed in Supplementary Table S1, along with country of origin, chromosome number, and the year they were imported into the USDA germplasm collection, if known. The leaves were air dried and DNA extracted using a modified CTAB method (Aitken et al. 2005). DNA was stored at − 20° C until required then subsampled and dried for transport. It was resuspended in 20 ul sH2O upon arrival in Panama.
Genetic analysis
Two noncoding chloroplast (cpDNA) gene regions (Regions no. 6 and 19 in Takahashi et al. 2005) were analyzed to identify cpDNA haplotypes in a subset of samples. Samples from the native range were chosen for sequencing based on geographic origin to explore haplotype variation across the range of S. spontaneum. All accessions from the World collection that were likely brought to Panama in 1939 were sequenced (see below). Similarly, samples from Panama were sequenced from throughout the invasive range, with a focus on the Panama Canal Watershed which is the likely origin of the invasion.
PCR reactions were carried out in 25 ul volumes containing 1 ul DNA, 1× PCR buffer, 25 mM MgCl2, 2 mM dNTPs, 2 uM of each primer, and 1 U AmpliTaq polymerase (Applied Biosystems, Inc., Carlsbad, CA). Reactions were performed on an ABI thermal cycler with the following cycling conditions: 94 °C for 3 min, 35 cycles of 94 °C for 1 min, 52 °C for 1 min, and 72 °C for 2 min, followed by a 7 min final extension at 72 °C. PCR reactions were cleaned using GELase enzyme (Epicentre), then sequenced using BigDye chemistry (Applied Biosystems) on an ABI 3100 automated sequencer. Sequences were aligned by eye using Sequencher 4.21 (GeneCodes) and insertion/deletion (indel) mutations were coded as single base characters to treat them as units rather than multiple character changes. Poly-A repeat regions were excluded from the analysis. Data from the two gene regions were combined to assign haplotype designations and a network was constructed using the statistical parsimony algorithm of Templeton and colleagues (Clement et al. 2000; Templeton et al. 1992) as implemented in popART (Leigh and Bryant 2015).
We assessed multilocus allele profiles in all samples (362 World collection, 186 Panama) using eight microsatellite loci (msscir14, msscir17, SMC221, SMC334, SMC336, SMC1047, SMC1237, SMC1493; Brown et al. 2007; Pan 2006) targeting different regions of the genome. PCR amplifications were performed in 7 µL reaction volumes containing approximately 10 ng DNA, 0.056 U Amplitaq Gold (Applied Biosystems, Inc., Carlsbad, CA), 1x reaction buffer, 3 mM MgCl2, 200 μM dNTPS, 0.5 pmol of forward primer including an M13 tag, 2.5 pmol of reverse primer, 2 pmol of FAM, PET, VIC, or NED labelled M13 primer, and 1 mg mL− 1 BSA. All PCR reactions were conducted with 2 min of denaturation at 94 °C, 35 cycles of denaturation at 94 °C for 45 s, annealing at 56 °C for 25 s, and 25 s of extension at 72 °C, and a final extension period of 72 °C for 5 min. Primers were not multiplexed due to the high ploidy level of the species, but PCR reactions with differing fluorescent tags were multiloaded, along with LIZ500 size standard, for capillary electrophoresis on an ABI Prism 3100 automated sequencer. Fragment sizes were analyzed using GeneMapper 3.7 (Applied Biosystems Inc.) and individuals were scored for presence or absence of identified alleles for each locus. As allele profiles were complex due to multiple ploidy levels across the dataset, we developed a scoring criteria to identify alleles to include in the final analyses. All scored alleles had consistent results over replicate amplifications and the maximum number of alleles scored per individual at a given locus was 10. As allele frequencies are unknown, multilocus allele profiles are hereafter referred to as allele phenotypes.
Data analysis
As sampling at the stand level was limited (e.g. only one ramet per stand at most sites), within-site measures of genetic diversity were not calculated and our analysis focuses on diversity within the S. spontaneum population in Panama compared to other locations. Replicate identical genotypes from samples collected along transects (e.g. clonal replicates; Bonnett et al. 2014) were removed from our Panama dataset prior to further analysis leaving us with 154 samples.
The R package poppr (Kamvar et al. 2014) was used to calculate the total number of alleles (A), and expected heterozygosity (HE) per locus for all samples from Panama (Panama), for all accessions in the modern germplasm collection from elsewhere in the world (World collection), and for the subset of accessions from the World collection that were likely present in the sugarcane germplasm collection brought to Panama in 1939 (Historic collection). In 1939 the collection likely included 25 accessions of S. spontaneum (Brandes et al. 1938), of which we had access to DNA from 12 accessions. We also included another nine accessions in our “Historic” group which could have been brought to Panama as they are recorded as having been imported to the US collection prior to 1939 (USDA-GRIN 2009; Table 1).
We characterized differences between geographic regions in several ways. First, genetic differentiation between populations was tested with AMOVA using the ploidy independent summary statistic ρ, as implemented in poppr. Information on groupings of samples can be found in Supplemental Table S1. Next, the R package polysat (Clark and Jasieniuk 2011) was used to generate matrices of distances between samples using the Lynch distance (Lynch 1990) which were then visualized using Principle Coordinates Analysis. Finally, we used Bayesian methods implemented in STRUCTURE 2.3.3 (Pritchard et al. 2000, Falush et al. 2007) to assign samples to clusters. STRUCTURE was run with an admixture model with correlated allele frequencies with 10 replicates for each run. Ploidy was set at 10 for all samples as the maximum number of alleles for any sample at a given locus was 10. Up to 20 genetic groups were simulated (K = 1–20) and each run consisted of a burn-in period of 10,000 MCMC (Monte Carlo Markov Chain) steps followed by 50,000 iterations. The most likely number of genetic clusters was determined with the Evanno et al. (2005) method, implemented in the software STRUCTURE HARVESTER v0.6.94 (Earl and vanHoldt 2012).
To estimate ploidy levels of samples from Panama, we compared the mean numbers of alleles per locus of samples from Panama (n = 154) with those from the World collection with known ploidy levels (n = 84; Table S1). These samples have reported chromosome counts ranging from 40 to 124 chromosomes.
Results
Chloroplast haplotype diversity
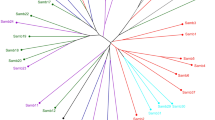
A total of nine haplotypes were identified, after combining sequences from two chloroplast gene regions, in 127 samples from the World collection and 54 from Panama (Fig. 1, Table S1). There were 30 segregating sites in the chloroplast sequence, of which 12 were parsimony informative. Haplotypes A-E were rare, with A, B and C found only in samples from Indonesia and India and D and E found in China and Taiwan. Haplotypes F-I had a broader distribution, ranging from east Africa to Papua New Guinea. Haplotype F was the most common (68% of samples sequenced from the native range) and had the broadest distribution across the native range of S. spontaneum. It was found in eastern Africa, the Middle East, India and other countries in South Asia, Indonesia, Papua New Guinea, China, and Taiwan (Table S1). It was also common in our subset of samples representing the Historic sugarcane germplasm collection that was brought to Panama in 1939 (75%; Table 1) and was the only haplotype found in modern S. spontaneum samples from Panama (100%). Other haplotypes found in the Historic collection were Haplotype H, found in three samples originating from Burma, India, and Papua New Guinea, and Haplotype I, found in one sample from Indonesia.
Haplotype network of Saccharum spontaneum cpDNA diversity obtained from sequencing 181 samples collected across the native and introduced ranges. Haplotypes were constructed by combining sequences from two loci (Regions no. 6 and 19 in Takahashi et al. 2005). Dashes between haplotypes represent mutations, following coding of indels as single characters. Unlabeled nodes indicate inferred steps not found in the sampled populations
Diversity of allele phenotypes
We genotyped 362 samples from the World collection and 154 from Panama with eight microsatellite markers that we could reliably score across all samples (Table 2). All samples from Panama displayed unique allele phenotypes. Similarly, all 362 accessions from the World collection had unique allele phenotypes except two, labelled “Formosa No.4” and “Gehra Bon”. It is not known what the correct identity of this clone is and whether the duplication resulted from an error in leaf sampling or during collection maintenance when germplasm was replanted, as the two accessions may have originally been planted side by side in alphabetical order in the USDA germplasm collection, leading to the potential for mislabeling. We included this allele phenotype in all analyses as a single sample of unknown origin as both of these accessions were present in the Historic germplasm collection and we did not want to eliminate a potential invasion source from our analysis.
Overall allelic diversity was extremely high and a total of 319 alleles were scored across the eight loci, with a mean number of alleles per locus (AO) of 39.6 (range 30–62; Table 2). Observed heterozygosity across all markers was nearly 100%, with a mean expected heterozygosity (HE) of 0.93 and evenness of 0.73. A subset of 177 alleles across all loci were found in samples from Panama, with a mean AO of 21.0 (range 14–29). Observed heterozygosity was also high in Panama, with a mean HE of 0.91 and evenness of 0.83 across loci. No private alleles were found in samples from Panama when compared with the World collection. Our subset of samples from the World collection representing accessions from the Historic collection (n = 21) had a mean of 22.1 alleles per locus (range 18–32) and HE was 0.91 and evenness was 0.81. Across all loci, 24 alleles were found in Historic samples that were not present in samples from Panama, and 18 alleles were found in Panama that were not in the Historic samples.
Regional structuring
Differences among plants originating from 16 different countries or regional groups (see Table S1 for grouping information) were tested using AMOVA. We found that 16.5% of the variance was present among groups whereas 85.5% was within groups (ϕPT=0.164), suggesting little clustering across groups.
Similarly, principle coordinates analysis (PCoA) showed a great deal of overlap between samples from different geographic regions in the native range, with little variability explained by each axis (Fig. 2a). No country or region formed a distinct cluster that did not overlap with other countries but a general West – East grouping of samples from India, Indonesia/Thailand, and Phillippines can be seen along PCoA axis 1.
Principle coordinates analysis plots based on pair-wise Lynch distances showing: a genetic differentiation in accessions from the World collection originating from across the native range of S. spontaneum; each circle represents a sample and sample sizes by country/region are indicated by numbers in parentheses in the legend; and b relationships between modern samples from Panama (open triangles), accessions from the World collection (grey circles), and accessions from the Historic USDA germplasm collection. Historic accessions are shown in color with haplotype F indicated by circles, haplotype H by squares, and haplotype I as a triangle. Country of origin of historical accessions are indicated in parentheses: BAN = Bangladesh, E = Egypt, IN = India, IND = Indonesia, PNG = Papua New Guinea, TUR = Turkmenistan. Unknown = Formosa No. 4/Gehra Bon allele phenotype. Numbers in parentheses along the axes indicate the percent of variation explained by each axis
Both PCoA and STRUCTURE analysis showed strong separation between samples from Panama and the native range (World; Figs. 2b, 3). When Panama was included in the PCoA, it clustered away from the native range along axis 1 and showed little spread along axis 2 (Fig. 2b, Supplemental Fig. S1a). When axis 2 was plotted against axis 3, Panama showed broad overlap with samples from across the native range but grouped most closely with samples from Indonesia (Fig. S1b). Similarly, STRUCTURE clearly inferred two populations (K = 2; Panama and World (native range)) with only a few samples showing an intermediate genotype (Fig. 3). All samples from Panama belonged to one population (membership probability 87.1–99.7%). Thirteen samples from the native range clustered with Panama (70.2–99.7% probability) and 16 were transitional between the two populations (30–70% probability of clustering with Panama). Of these 29 samples, seven were found in the Historic collection (Djatiroto, Glagah_WT, Kloet, Kepandjen, Pasoeroean, Tabongo, and Gehra Bon/Formosa No.4). These same seven historical accessions also clustered with Panama in the PCoA and have high ploidy levels and Haplotype F (Table 1).
STRUCTURE plot from Saccharum spontaneum samples from the native (World) and introduced (Panama) ranges. Each bar represents a sample and colors show the assignment probabilities to inferred evolutionary clusters (K = 2) based on eight microsatellite loci. Historic refers to the subset of samples from the World collection that may have been brought to Panama in 1939
When compared to accessions that were likely in the Historic germplasm collection, the PCoA showed samples from Panama clustering primarily with accessions from Indonesia (eight of nine accessions overlapping with the Panama cluster, Fig. 2b). Other accessions clustering with Panama include the allele phenotype of ambiguous origin, produced by the samples Formosa No.4 (from Taiwan) and Gehra Bon (from India), and the selfed offspring of an accession from Turkmenistan (US4515; Fig. 2b). No historical accessions with cpDNA haplotypes H or I clustered with samples from Panama (Fig. 2b).
Ploidy
Our dataset of samples from the native range included 84 samples of known ploidy level (Supplemental Table S1), which have chromosome counts ranging from 2 N = 40–124 (likely 5X – 15X). Of these samples, 33 had Haplotype F which does not appear to be linked to any individual ploidy level as it was found in samples with chromosome counts ranging from 2 N = 40–112 that originate from throughout the native range. While we have no direct measure of the ploidy level of samples from Panama, the high observed allelic diversity across loci suggests that it is high (up to 10 alleles observed within a sample per locus). Although the total number of observed alleles across loci is lower in Panama than in the World collection (Table 2), the mean number of alleles per sample across all loci (5.3) is higher than in any other group of samples of known ploidy levels (Table 3; range 4.3–4.9). The Historic accessions that cluster with Panama in our PCoA and STRUCTURE analyses all had cpDNA haplotype F, high ploidy (2 N = 96–112), and a similar mean number of alleles per sample across loci (Tables 1, 3).
Discussion
The historical record indicates that the spread of S. spontaneum in Panama began within a decade of the arrival of the USDA sugarcane germplasm collection at the CZEG in 1939 (Moore 1948). Although we did not have access to all of the accessions in the historical USDA collection (likely 25–35 accessions; Artschwager 1942; Brandes et al. 1938; Price 1957; USDA-GRIN 2009), our Historic dataset includes 21 accessions and is representative of both the geographic range and variation in ploidy of specimens in the germplasm collection at the time (Table 1). Today the S. spontaneum population in Panama represents a single cpDNA haplotype lineage and is genotypically distinct from the majority of samples in the world S. spontaneum germplasm collection which shows high genetic diversity and little geographic structuring (Fig. 2a, b). This single haplotype is the most common haplotype across the native range of the species and the most common in S. spontaneum accessions brought to Panama in 1939 in the USDA collection (Table 1). Although heterozygosity levels at all microsatellite loci were elevated due to high ploidy levels, they were reduced in samples from Panama compared to accessions from elsewhere in the world and the majority of microsatellite alleles found in Panama today were also found in the historical USDA accessions that we have tested. Further, all accessions from the Historic collection that clustered with Panama in our analyses had the same cpDNA haplotype and similar numbers of alleles/locus suggesting similar ploidy levels in both groups. This provides convincing evidence for the escape of S. spontaneum from the germplasm collection into natural settings in Panama, most likely in the early 1940’s when growth experiments, which allowed plants to flower and set seed, were being conducted at the CZEG (Brandes 1950).
Genetic diversity and reproduction
Genetic diversity of S. spontaneum across the landscape in Panama is extremely high with high levels of heterozygosity and duplicate genotypes found only in samples collected immediately adjacent to each other (this study; Bonnett et al. 2014). This is likely due to its generalist reproductive strategy, where both pollen and seeds are wind dispersed, promoting high outcrossing rates and seed dispersal which helps to maintain allelic richness and maximizes genetic variation in propagules as they spread to new locations (Richardson and Pyšek 2012; Williamson 2006). However, S. spontaneum also has the ability to self-fertilize (Panje 1970), ensuring that viable seed can be produced and dispersed annually even in isolated stands. Both seed production and germinability are also high (Bonnett et al. 2014), and as ample high-light environments exist across the landscape, establishment of new stands occurs annually and growth is rapid once plants establish (K. Saltonstall, pers. obs). Further, the high ploidy levels of S. spontaneum do not appear to impact its ability to produce viable seed and have likely helped it to maintain high allelic diversity in both its native and introduced ranges (te Beest et al. 2012).
We find it extremely likely that the establishment of S. spontaneum in Panama was facilitated by intraspecific hybridization of multiple genetically distinct accessions. This has been seen with other invaders such as reed canary grass (Phalaris arundinacea L.) where repeated introductions of accessions native to different parts of Europe has alleviated inbreeding depression and allowed for rapid evolution to occur in North America (Lavergne and Molofsky 2007). The population in Panama does show an overall reduced number of alleles compared to samples from across the native range of S. spontaneum which could be the result of a founder effect or inbreeding depression. However, 177 alleles were found in only eight microsatellite loci, all of which are found in accessions from the World collection. In addition, in comparison to the Historic subset of 21 accessions, there were several alleles found in samples from Panama that were not in the Historic dataset. As our dataset is incomplete in that we did not have access to DNA from all accessions that could have been brought here in 1939, it is plausible that one or more of the missing accessions could have contributed genetic material to the founding propagules of this invasion.
Further, given that seven of the nine samples from our Historic group that cluster with Panama in both our cpDNA haplotype sequencing (Fig. 1) and nuclear microsatellite analyses (Figs. 2c, 3,) originate from Indonesia, it is plausible that accessions from Indonesia were the primary sources of propagules of this invasion, either via pollen or seed dispersal. When plotted, axes 2 and 3 of the PCoA of all samples also support this conclusion as samples from Panama cluster most closely with those from Indonesia (Supp. Figure 1b). Indonesia is located along the equator and has a similar light regime and tropical climate to Panama, with seasonality of rainfall and consistently high temperature and humidity levels.
The role of pre-adaptation in invasion success
The success of the S. spontaneum invasion in Panama may also reflect the consequences of pre-adapted genotypes or broad environmental tolerance (e.g. phenotypic plasticity) or a combination of both. We suggest that pre-adaptation for various life-history and physiological traits ensured high fitness and success of this invader in Panama and highlight the following traits:
Firstly, although the ploidy level of S. spontaneum in Panama is unknown, allele phenotype patterns suggest that it is high (likely chromosome count of 2 N = 96+, Table 3) ensuring that the founding propagules of this invasion had high genetic diversity across their genomes. Genetic diversity remains high, even after 70–80 years of isolation from other S. spontaneum populations. Observed heterozygosity is also high across loci, although lower than that seen in the modern World collection. This likely reflects the isolation of this population and a founder effect, but the high ploidy level has helped to counteract the forces of genetic drift by providing genetic redundancy and possibly helping to mask the effects of deleterious alleles (te Beest et al. 2012 and references therein).
Second, the mixed breeding strategy and high reproductive output of S. spontaneum (Bonnett et al. 2014) would have facilitated a small number of propagules to disperse, establish, and then spread into the new environment. While the preference for outcrossing helps to maintain genetic diversity and reduce inbreeding depression, the ability to self gives reproductive assurance along the leading edge of an invasion where stands may be isolated (Novak and Mack 2005). Traits such as small seed size and high dispersal capability via wind, as seen in many invasive grasses, have also been shown to facilitate invasions (Rejmanek and Richardson 1996).
Third, as a C4 perennial plant, S. spontaneum has high water and nitrogen use efficiency allowing it to grow well under elevated temperatures (Cernusak et al. 2007). Its high ploidy level may also contribute to its tolerance to dry periods, as polyploids typically have larger and fewer stomata, resulting in lower transpiration rates and reduced water loss than diploids (Li et al. 1996). As such, it can persist and maintain photosynthetic tissues throughout the year, including during the three to four month dry season of central Panama when many other plants go dormant (K. Saltonstall, pers. observation). Further, the increased prevalence of dry season fires in S. spontaneum-dominated parts of the PCW serve to stimulate its growth and reproduction, giving it a competitive advantage over native plants that are killed by fire (Saltonstall and Bonnett 2012).
Finally, previous work has shown that S. spontaneum is sensitive to photoperiod (Brandes 1950; Panje and Srinivasan 1959; Sartoris 1939) and needs high light levels to maximize its growth. In a growth study which included seven S. spontaneum accessions grown at six different locations (3 tropical, 3 temperate), Brandes (1950) found that accessions originating from lower latitudes tended to grow better at CZEG than the other two tropical sites (Puerto Rico and Colombia). At 9° N latitude, day length in Panama varies little over the course of the year and in open areas, such as pastures and agricultural lands where S. spontaneum frequently invades, light levels are high year round. These conditions, in combination with the seasonality of rainfall, are ideal for the growth of S. spontaneum (Brandes 1950; Panje and Srinivasan 1959). Although historically dominated by moist tropical rainforest, the area around the CZEG (today Summit Gardens in Soberania National Park) in the PCW was largely deforested during the construction of the Panama Canal and subsequent expansion of agriculture in the region (Fig. 4). Accidental dispersal and establishment of seeds from the germplasm collection is entirely plausible as there were growth experiments happening at the time that allowed plants to flower, at least for the seven accessions included in the experiment, all of which we are fortunate to have analyzed in this study (Table 1; Brandes 1950). Saccharum spontaneum seeds germinate readily on disturbed and exposed soils where light levels are high and soil moisture is adequate (Bonnett et al. 2014) and this type of environment surrounded the CZEG in 1939. Once established, growth rates are high when moisture is adequate and plants can begin flowering in the first year of growth (Brandes 1950; K. Saltonstall, unpub. data).
Aerial view of the CZEG (then called the Canal Zone Plant Introduction Gardens) in 1923 (CZPIG 1929)
Biosecurity of germplasm collections
The invasion of S. spontaneum in Panama represents a “perfect storm” of circumstances that allowed this plant to establish and spread rapidly throughout the PCW and demonstrates the importance of careful management of germplasm collections. A diverse collection of sugarcane germplasm, that included many accessions from tropical regions, was deliberately brought to Panama and maintained under conditions to ensure their growth and flowering. High genetic diversity due to high ploidy levels coupled with pre-adaptation to the local conditions then gave it high reproductive and competitive success in a disturbed tropical environment. Intraspecific hybridization between accessions may have also contributed to its success. Once established, it was able to outcompete other native and introduced grasses and quickly spread across the region.
We emphasize however that this invasion represents an accidental escape from a germplasm collection which was brought to Panama for the best of intentions. Invasion potential was not considered as a risk factor in the 1930’s when transporting plants for agricultural purposes and the focus of the sugarcane collection was crop improvement and germplasm preservation (CZEG 1939). Experimental work at the time emphasized understanding of growth and reproductive patterns under different environmental conditions, such as rainfall and photo1939period (Brandes 1950). Today there are strict rules in place to prevent accidental escape from germplasm collections (e.g. prevention of flowering, use of concrete pads under pots to prevent rooting in the soil and vegetative spread, Bretting 2007; FAO 2013), but accidents will happen and there is always the potential for escape to occur, particularly with taxa that are known to exhibit invasive spread in other parts of the world (Westbrooks and Miller 1993). Continuous monitoring and rapid response to accidental escape and establishment of potential invaders are important management tools that cannot be minimized or overlooked, particularly given current scenarios of global change. Many millions of dollars have been spent in Panama on control of S. spontaneum as efforts to reforest the PCW have been a priority of the Panama Canal Authority (Craven et al. 2009; MGM Innova 2010; Wishnie et al. 2002) yet it continues to expand its distribution and impede reforestation efforts (Boeschoten et al. 2020). We hope that this example serves as a reminder of unintended consequences despite the best intentions of researchers and managers alike.
Data accessibility
Sequences reported in this study have been deposited in the GenBank database (Accessions MT978044-978050, MT982603-MT982608). The microsatellite datasets generated from this study are available on request from the corresponding author.
References
Ahmad R, Liow P-S, Spencer DF, Jasieniuk M (2008) Molecular evidence for a single genetic clone of invasive Arundo donax in the United States. Aquat Bot 88:113–120
Aitken KS, Jackson PA, McIntyre CL (2005) A combination of AFLP and SSR markers provides extensive map coverage and identification of homo(eo)logous linkage groups in a sugarcane cultivar. Theor Appl Gen 110:789–801
Aitken KS, Li J, Piperidis G, Qing C, Yuanhong F, Jackson P (2018) Worldwide genetic diversity of the wild species and level of diversity captured within sugarcane breeding programs. Crop Sci 58:218
ANAM-ACP (2006) Report: Monitoreo de la Cuenca Hidrográfica del Canal de Panamá. Panama
Artschwager E (1942) A comparative analysis of the vegetative characteristics of some variants of Saccharum spontaneum. USDA Tech Bull No. 811. Washington, DC: United States Department of Agriculture
Boeschoten LE, van Breugel M, Bailon M, Balbuena J, Nuñez M, Cerezo A, Hall JS (2020) Framework species approach proves robust in restoring forest on fire prone invasive grass: a case study from Panama. J Sust For. https://doi.org/10.1080/10549811.2020.1746915
Bonnett GD, Kushner JNS, Saltonstall K (2014) The reproductive biology of Saccharum spontaneum L.: implications for management of this invasive weed in Panama. NeoBiota 20:61–79
Brandes EW (1950) Changes in seasonal growth gradients of geographically displaced sugar cane. Proc Cong Int Soc Sugarcane Tech 7:1–32
Brandes EW, Sartoris GB (1936) Sugar cane: its origin and improvement. In: Yearbook of agricultural research. US Department of Agriculture, pp. 561–623
Brandes FW, Sartoris GB, Grassl CO (1938) Assembling and evaluating wild forms of sugarcane and closely related plants. Proc Cong Int Soc Sugarcane Tech 6:128–154
Bretting PK (2007) The US National Plant Germplasm System in an era of shifting international norms for germplasm exchange. Acta Hort 760:55–60
Brown J, Schnell R, Power E, Douglas S, Kuhn D (2007) Analysis of clonal germplasm from five Saccharum species: S. barberi, S. robustum, S. officinarum, S. sinense and S. spontaneum. A study of inter-and intraspecies relationships using microsatellite markers. Gen Res Crop Evol 54: 627–648
Canavan S, Meyerson LA, Packer JG, Pyšek P, Maurel N, Lozano V,…Wilson JRU (2019) Tall-statured grasses: a useful functional group for invasion science. Biol Invasions 21: 37–58
Cerezo A (2010) Antecedentes del origen y objetivo de la introducción de la maleza paja blanca (Saccharum spontaneum L.) a Panamá. Panama: Autoridad del Canal de Panamá. Departamento de Ambiente, Agua y Energía.http://www.cich.org/publicaciones/05/paja-blanca.pdf. Accessed 28 May 2019
Cernusak L, Aranda J, Marshall JD, Winter K (2007) Large variation in whole-plant water-use efficiency among tropical tree species. New Phytol 173:294–305
Clark LV, Jasieniuk M (2011) POLYSAT: an R package for polyploid microsatellite analysis. Mol Ecol Res 11:562–566
Clement M, Posada D, Crandall KA (2000) TCS: a computer program to estimate gene genealogies. Mol Ecol 9:1657–1659
Condit R, Robinson WD, Ibáñez R, Aguilar S, Sanjur A, Martínez R, … Heckadon S (2001) The status of the Panama Canal Watershed and its biodiversity at the beginning of the 21st century. Bioscience 51:389–398
Cordeiro GM, Taylor GO, Henry RJ (2000) Characterization of microsatellite markers from sugarcane (Saccharum sp.), a highly polyploid species. Plant Sci 155:161–168
Craven D, Hall JS, Verjans J-M (2009) Impacts of herbicide application and mechanical cleaning on growth and mortality of two timber species in Saccharum spontaneum grasslands of the Panama Canal Watershed. Restor Ecol 17:751–761
Croat TB (1971) Summit Garden, Panama Canal Zone: its role in botanical research. Taxon 20:769–772
CZEG (1939) Annual Report of the Canal Zone Experiment Gardens for the Fiscal Year 1939 (28 pgs). Mount Hope, Canal Zone Panama
CZPIG (1929) Annual Report of the Canal Zone Plant Introduction Gardens for the Fiscal Year 1928 (43 pgs). Mount Hope, Canal Zone Panama
D’Antonio C, Vitousek PM (1992) Biological invasions by exotic grasses. Ann Rev Ecol Syst 23:63–87
D’Antonio C, Stahlberger K, Molinari N (2011) Grasses and forbs. In: Simberloff D, Rejmanek M (eds) Encyclopedia of biological invasions. University of California Press, Berkeley, pp 280–290
Dlugosch KM, Anderson SR, Braasch J, Cang FA, Gillette HD (2015) The devil is in the details: genetic variation in introduced populations and its contribution to invasion. Mol Ecol 24:2095–2111
Dlugosch KM, Parker IM (2008) Founding events in species invasions: genetic variation, adaptive evolution, and the role of multiple introductions. Mol Ecol 17:431–449
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Con Gen Resour 4:359–361
Ellstrand NC, Schierenbeck KA (2000) Hybridization as a stimulus for the evolution of invasiveness in plants? Proc Nat Acad Sci 97:7043–7050
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Falush D, Stephens M, Pritchard JK (2007) Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol 7:574–578
FAO (2013) Genebank standards for plant genetic resources for food and agriculture. FAO, Rome
Gioria M, Pyšek P (2017) Early bird catches the worm: germination as a critical step in plant invasion. Biol Invasions 19:1055–1080
Hammond B (1999) Saccharum spontaneum (Gramineae) in Panama: the physiology and ecology of invasion. J Sust For 8:23–28
Hardion L, Verlaque R, Saltonstall K, Leriche A, Vila B (2014) Origin of the invasive Arundo donax (Poaceae): a trans-Asian expedition in herbaria. Ann Bot 114:455–462
Heckadon-Moreno S, Ibáñez R, Condit R (1999) La cuenca del Canal: Deforestación, contaminación y unbanización [Executive Summary]. Instituto Smithsonian de Investigaciones Tropicales, Panama
Hollingsworth ML, Bailey JP (2000) Evidence for massive clonal growth in the invasive weed Fallopia japonica (Japanese Knotweed). Bot J Linn Soc 133:463–472
Holm LG, Plucknett DL, Pancho JV, Herberger JP (1977) Saccharum spontaneum L. In: World weeds: natural histories and distribution. University of Hawaii Press, Honolulu, pp 693–698
Hooper E, Legendre P, Condit R (2005) Barriers to forest regeneration of deforested and abandoned land in Panama. J Appl Ecol 42(6):1165–1174
Hurka H, Bleeker W, Neuffer B (2003) Evolutionary processes associated with biological invasions in the Brassicaceae. Biol Invasions 5:291–292
Jones ER, Wishnie MH, Deago J, Sautu A, Cerezo A (2004) Facilitating natural regeneration in Saccharum spontaneum (L.) grasslands within the Panama Canal Watershed: effects of tree species and tree structure on vegetation recruitment patterns. For Ecol Manag 191:171–183
Kamvar Z, Tabima J, Grünwald N (2014) Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2:e281
Lambert AM, Dudley TL, Saltonstall K (2014) Ecology and impacts of the large-statured invasive grasses Arundo donax and Phragmites australis in North America. Invasion Plant Sci Manag 3:489–494
Lavergne S, Molofsky J (2007) Increased genetic variation and evolutionary potential drive the success of an invasive grass. Proc Nat Acad Sci 104:3883–3888
Le Roux JJ, Wieczorek AM, Wright MG, Tran CT (2007) Super-genotype: global monoclonality defies the odds of nature. PLoSOne 7:e590
Leigh J, Bryant D (2015) PopART: full-feature software for haplotype network construction. Methods Ecol Evol 6:1110–1116
Li W-L, Berlyn GP, Ashton PMS (1996) Polyploids and their structural and physiological characteristics relative to water deficit in Betula papyrifera. Am J Bot 83:15–20
Lynch M (1990) The similarity index and DNA fingerprinting. Mol Biol Evol 7:478–484
MGM Innova (2010) Panama Canal Authority sustainable forest cover establishment project: project design document for Climate, Community, & Biodiversity Standards (CCBS) Project Design Document, 2nd edn. Panama City, Panama, p 146
Moore GT (1948) Fifty-ninth annual report of the Director. MO Bot Gard Bull 36(1):16
Nepstad DC, Uhl C, Serrão EAS (1991) Recuperation of a degraded Amazonian landscape: forest recovery and agricultural restoration. Ambio 20:248–255
Novak SJ, Mack RN (2005) Genetic Bottlenecks in alien plant species: Influence of mating systems and introduction dynamics. In: Sax DF, Stachowicz JJ, Gaines SD (eds) Species invasions: insights into ecology, evolution, and biogeography. Sinauer Associates, Inc, Sunderland, pp 202–228
Pan Y-B (2006) Highly polymorphic microsatellite DNA markers for sugarcane germplasm evaluation and variety identity testing. Sugar Tech 8:246–256
Pandit M, Pocock MJO, Kunin WE (2011) Ploidy influences rarity and invasiveness in plants. J Ecol 99:1108–1115
Panje RR, Babu CN (1960) Studies in Saccharum spontaneum: distribution and geographical association of chromosome numbers. Cytologia 25:152–172
Panje RR, Srinivasan K (1959) Studies in Saccharum spontaneum. The flowering behavior of latitudinally displaced populations. Bot Gaz 120:193–202
Panje RR (1970) The evolution of a weed. Trop Pest Manag 16:590–595
Parker IM, Gilbert GS (2007) When there is no escape: the effects of natural enemies on native, invasive, and noninvasive plants. Ecology 88:1210–1224
Parsons JJ (1972) Spread of African pasture grasses to the American tropics. J Range Manag 25:12–17
Price S (1957) Cytological studies in Saccharum and allied genera. III Chromosome numbers in interspecific hybrids. Bot Gaz 118:146–159
Price S (1965) Cytology of Saccharum robustum and related sympatric species and natural hybrids. Tech Bull No. 1337. Washington, DC: USDA Agricultural Research Service
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Rejmanek M, Richardson DM (1996) What attributes make some plant species more invasive? Ecology 77:1655–1661
Richards CL, Bossdorf O, Muth NM, Gurevitch J, Pigliucci M (2006) Jack of all trades, master of some? On the role of phenotypic plasticity in plant invasions. Ecol Lett 9:981–993
Richardson DM, Pyšek P (2012) Naturalization of introduced plants: ecological drivers of biogeographical patterns. New Phytol 196:383–396
Sakai AK, Allendorf FW, Holt JS, Lodge DM, Molofsky J, With KA, … Weller SG (2001) The population biology of invasive species. Ann Rev Ecol Syst 32:305–332
Saltonstall K, Bonnett GD (2012) Fire promotes growth and reproduction of Saccharum spontaneum (L.) in Panama. Biol Invasions 14:2479–2488
Sartoris GB (1939) The behavior of sugar cane in relation to length of day. Proc Cong Int Soc Sugarcane Tech 6:796–801
Soltis PS, Soltis DE (2000) The role of genetic and genomic attributes in the success of polyploids. Proc Nat Acad Sci USA 97:7051–7057
Takahashi S, Furukawa T, Asano T, Terajima Y, Shimada H, Sugimoto A, Kadowaki K (2005) Very close relationship of the chloroplast genomes among Saccharum species. Theor Appl Genet 110:1523–1529
te Beest M, Roux L, Richardson JJ, Brysting DM, Suda AK, Kubešová J, M., & Pyšek P (2012) The more the better? The role of polyploidy in facilitating plant invasions. Ann Bot 109:19–45
Templeton A, Crandall KA, Sing CF (1992) A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics 132:619–633
USDA (2016) Weed Risk Assessment for Saccharum spontaneum L. (Poaceae)—Wild sugarcane. USDA APHIS. https://www.aphis.usda.gov/plant_health/plant_pest_info/weeds/downloads/wra/Saccharum-spontaneum.pdf. Accessed 15 May 2019
USDA-GRIN (2009) Saccharum spontaneum accessions. https://npgsweb.ars-grin.gov/gringlobal/search.aspx. Accessed 24 Apr 2009
von Lindeman G (1986) Origen, establecimiento y problemas potenciales de la maleza Saccharum spontaneum en Panama. Centro Agronómico Tropical de Investigación y Enseñanza, Turrialba
Westbrooks RG, Miller JD (1993) Investigation of wild sugarcane (Saccharum spontaneum L.) escaped from the USDA ARS sugarcane field station at Canal Point, Florida. Proc South Weed Sci Soc 46:293–295
Williams DG, Baruch Z (2000) African grass invasion in the Americas: Ecosystem consequences and the role of ecophysiology. Biol Invasions 2:123–140
Williamson M (2006) Explaining and predicting the success of invading species at different stages of invasion. Biol Invasions 8:1561–1568
Wishnie MH, Deago J, Mariscal E, Sautu A (2002) The efficient control of Saccharum spontaneum (L.) (Graminae) in mixed plantations of six native species of tree and teak (Tectona grandis) in the Panama Canal Watershed, Republic of Panama (No. ECO-03-03-En, p. 34. Smithsonian Tropical Research Institute, Panama City
Acknowledgements
Many thanks to Dr. Eldredge Bermingham for the use of his laboratory at STRI and his support to KS. All samples from Panama were collected under ANAM permit No. SIM/P-1-09. KS was supported by a Smithsonian Postdoctoral Fellowship. GDB was supported by the Queensland government by a Queensland-Smithsonian Research Fellowship, and KA and GDB were supported by the Australian Government through the Co-operative Research Centre for Sugar Industry Innovation through Biotechnology and the Sugar Research and Development Corporation (now Sugar Research Australia). We thank Jing-Chuan Li for sharing DNA extractions from the world collection. We also thank two anonymous reviewers for their thoughtful comments and suggestions.
Author information
Authors and Affiliations
Contributions
KS designed the research, performed the lab work, analyzed the data, and wrote the manuscript. GB assisted with field collections and KA provided DNA from the sugarcane germplasm collection. All authors commented on drafts of the manuscript and have read and approved the final manuscript.
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Saltonstall, K., Bonnett, G.D. & Aitken, K.S. A perfect storm: ploidy and preadaptation facilitate Saccharum spontaneum escape and invasion in the Republic of Panama. Biol Invasions 23, 1101–1115 (2021). https://doi.org/10.1007/s10530-020-02421-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10530-020-02421-3