Abstract
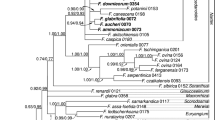
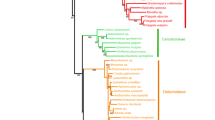
During the past three decades, molecular taxonomy has made considerable changes in the systematic delimitations of several families in the order Ericales which were formed earlier based on morphology. For instance, the Pentaphylacaceae s.l. has been treated differently by both modern and traditional taxonomists. Modern molecular taxonomists constituted this family by combining the traditionally defined Pentaphylacaceae s.s. (Pentaphylax), Sladeniaceae s.s. (Sladenia), the subfamily Ternstroemioideae with 11 genera of Theaceae s.l. and the genus Ficalhoa. There are also treatments placing the genus Pentaphylax with Ternstroemioideae in Pentaphylacaceae and Ficalhoa with Sladenia in Sladeniaceae. Because most of these genera are poorly studied, investigations on all aspects are important to understand the phylogeny to settle the issues surrounding the treatment of the 14 genera in this family. In the present study, DNA sequences of nrITS and trnL-F genes from species of 11 genera from these 14 genera were generated and analyzed together with sequences from other closely related members of Ericales. The results suggested existence of four distinct lineages viz., Sladenia, Pentaphylax, and tribes Frezierieae (9 genera) and Ternstroemieae (2 genera). Further, it demonstrated that within the biggest lineage, Frezierieae, the Visnea remained sister to the clades Adinandra+Cleyera, Euryodendron+Symplococarpon, Freziera, and finally, Eurya. Based on the evidence, it can be concluded that Sladeniaceae and Pentaphylacaceae are very close to each other and the proposal of merging them into a mega family Pentaphylacaceae s.l. with four tribes, i.e., Sladenieae, Pentaphylaceae, Ternstroemieae, and Freziereae should be considered seriously.


Similar content being viewed by others
References
Akaike H (1974) A new look at the statistical model identification. IEEE Trans Autom Control 19:716–723
Anderberg AA, Rydin C, Kallersjo M (2002) Phylogentic relationships in the order Ericales s.l.: analyses of molecular data from five genes from the plastid and mitochondrial genomes. Am J Bot 89:677–687
APG III (2009) An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Bot J Linn Soc 161:105–121
Bajpai PK, Warghat AR, Sharma RK, Yadav A, Thakur AK, Srivastava RB, Stobdan T (2014) Structure and genetic diversity of natural populations of Morus alba in the Trans-Himalayan Ladakh Region. Biochem Genet 52(3):137–152
Cronquist A (1981) An integrated system classification of flowering plants. Columbia University Press, New York, USA
Cunningham CW (1997) Can three incongruence tests predict when data should be combined? Mol Biol Evol 14:733–740
Degnan JH, Rosenberg NA (2006) Discordance of species trees with their most likely gene trees. PLoS Genet 2:e68
Farris JS (1989) The retention index and the rescaled consistency index. Cladistics 5:417–419
Farris JS, Albert VA, Kallersjo M, Lipscomb D, Kluge AG (1996) Parsimony jackknifing outperforms neighbor-joining. Cladistics 12:99–124
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Feng SG, Lu JJ, Gao L, Liu JJ, Wang HZ (2014) Molecular phylogeny analysis and species identification of Dendrobium (Orchidaceae) in China. Biochem Genet 52(3):127–136
Fitch WM, Ye J (1991) Weighted parsimony: does it work? In: Miyamoto MM, Cracraft J (eds) Phylogenetic analysis of DNA sequences. Oxford Press, New York, USA, pp 147–154
Genetic Computer Group (1994) Program manual for the Wisconsin package, version-8. Genetic Computer Group, Madison, WI, USA
Hall TA (1999) Bioedit: a user friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucl Acid Symp Ser 41:95–98
Huelsenbeck JP, Ronquist FR (2001) MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755
Keng H (1962) Comparative morphological studies of in Theaceae. Univ Calif Publ Ser Bot 33:269–384
Kluge AG, Farris JS (1969) Quantitative phyletics and the evolution of anurans. Syst Zool 18:1–32
Kubitzki K (2004) The families and genera of vascular plants. VI. Flowering plants. Dicotyledons. Celastrales, oxalidales, rosales, cornales, ericales. Springer, Berlin, NY
Li L, Liang HX, Peng H, Lei LG (2003) Sporogenesis and gametogenesis in Sladenia and their systematic implication. Bot J Linn Soc 143:305–314
Luna VI, Ochoterena H (2004) Phylogenetic relationships of the genera of Theaceae based on morphology. Cladistics 20:223–270
Melchior H (1925) Theaceae. In: Engler HGA, Prantl K (eds) Die Natürlichen Pflanzenfamilien. II, vol 21. W. Englemann, Leipzig, Germany, pp 109–154
Merrill ED (1918) Notes on the floral of Loh Fau Mountain, Kwangtung Province, China. Philipp J Sci 13C:123–161
Posada D, Crandall KA (1998) MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817–818
Schönenberger J, Anderberg AA, Sytsma K (2005) Molecular phylogenetic and pattern of floral evolution in the Ericales. Int J Plant Sci 166:265–288
Simmons MP (2004) Independence of alignment and tree search. Mol Phylogenet Evol 31:874–879
Stevens P, Weitzman A (2004) Sladeniaceae. In: Kubitzki K (ed) The families and genera of vascular plants. VI. Flowering plants. Dicotyledons. Celastrales, oxalidales, rosales, cornales, ericales. Springer, Berlin, NY, pp 431–433
Swofford DL (2003) PAUP: Phylogenetic Analysis Using Parsimony (*and other methods). Version 4.0b10. Sinauer Associates, Sunderland, MA, USA
Takhtajan A (1997) Diversity and classification of flowering plants. Columbia University Press, New York, NY, USA
Thorne RF (1992) Classification and geography of the flowering plants. Bot Rev 58:225–350
Tsou CH (1995) Embryology of theaceae. Vol. 71—Anther and ovule development of Adinandra, Cleyera and Eurya. J Plant Res 108:77–86
Vijayan K, Tsou CH (2008) Phylogenetic implications of nrITS on Camellia: The need of high quality DNA and PCR amplification with Pfu-DNA polymerase. Bot Stud 49:177–188
Weitzman AL (1987) Systematics of Freziera wild (Theaceae). Ph.D. thesis, Department of Organismic and Evolutionary Biology, Harvard University, Boston, MA, USA
Weitzman AL, Dressler S, Stevens PF (2004) Ternstroemiaceae. In: Kubitzki K (ed) The families and genera of vascular plants. VI. Flowering plants. Dicotyledons. Celastrales, oxalidales, rosales, cornales, ericales. Springer, Berlin, NY, pp 450–460
Wu CC, Hsu ZF, Tsou CH (2007) Phylogeny and taxonomy of Eurya (Ternstroemiaceae) from Taiwan, as inferred from ITS sequence data. Bot Stud 48:97–116
Yang Z, Rannala B (1997) Bayesian phylogenetic inference using DNA sequences: a Markov Chain Monte Carlo method. Mol Biol Evol 14:717–724
Acknowledgments
The authors thank Arne Anderberg, Bruce Bartholomew, Jens Bittner, Dawn Frame, James Maxwell, Hua Peng, Hong Wang, Hua-Gu Ye, and Kwok L. Yip for help with field collections and/or providing samples; the curators of the herbaria of CAS, GH, NY, and S for providing specimens; the anonymous reviewers for giving useful comments; the National Science Council, Republic of China (Grant NSC 96-2312-B-001-012) and Academia Sinica, Republic of China, for financial support.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Tsou, Ch., Li, L. & Vijayan, K. The Intra-familial Relationships of Pentaphylacaceae s.l. as Revealed by DNA Sequence Analysis. Biochem Genet 54, 270–282 (2016). https://doi.org/10.1007/s10528-016-9717-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-016-9717-1