Abstract
Population genetics has been recognized as a key component of policy development for fisheries and conservation management and aquaculture development. This study aims to evaluate the genetic diversity and population structure of native cobia (Rachycentron canadum) in the Gulf of Thailand and Andaman Sea, establishing the existing population distributions and contributing information to aid in the development of policy, prior to extensive aquaculture development. Microsatellite analysis of natural cobia populations in these two ocean basins shows similar levels of gene diversity at 0.844 and 0.837, respectively. All populations and almost all microsatellite loci studied show significant heterozygote deficiency. Genetic differentiation between local populations is low and mostly not significant (R ST = −0.0109 to 0.0066). The population shows no marked structure over the long geographic barrier of the Thai–Malay peninsula, either when analyzed using pairwise genetic differences or evaluated without predefined populations using STRUCTURE. Additionally, a Mantel test shows no evidence of isolation by distance between the population samples. The significant heterozygote deficiency at most of the loci studied could be explained by the possibility of null alleles. Alternatively, given the behavior of forming small spawning aggregations, seasonal migration, and hitchhiking on large marine animals, the population genetics could be complex. The population of cobia at each location in Thai waters may be inbred, as a result of breeding between relatives, which would reduce heterozygosity relative to Hardy–Weinberg frequencies, while some of these populations could be making long distance migrations followed by admixture between resident and transient groups. This migration would cause population homogeneity in allele frequencies on a larger geographic scale. The results suggest that fisheries management for this species should be considered at both national and international levels, and until the possibility of local adaptation is fully investigated, policy development should apply the precautionary principle to ensure the preservation of genetic diversity and the sustainability of local and regional fisheries.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Knowledge of the genetic diversity and population structure of wild stocks can aid policy design with regard to fisheries and conservation management, as well as aquaculture development (Hutchings and Fraser 2008; Kuparinen and Merilä 2007; Palumbi 2003). Information on genetic diversity of wild populations is also an essential component in the start of any breeding program for developing domesticated stocks for farm culture (Fjalestad et al. 1993; Gjøen and Bentsen 1997; Hena et al. 2005; Rezk et al. 2003). It has been shown that overexploitation in fisheries can induce a rapid loss of genetic variation of target species, which may diminish the adaptability and persistence of the species (Hauser et al. 2002; Hutchinson et al. 2003; Smith et al. 1991; Turner et al. 2002). A lack of a policy regarding farming industries can result in introgression of non-native or domesticated genes into wild populations, potentially causing a loss in genetic variability (Bekkevold et al. 2006; Fleming et al. 2000; Hindar et al. 2006; McGinnity et al. 2003). Investigating genetic diversity and population structure of species, after overexploitation and/or introgression have occurred, fails to maximize the potential for using this knowledge as a tool to aid in the development of policy which adequately protects biodiversity in the wild populations from anthropogenic influences. This study aims to evaluate the genetic diversity and population structure of wild cobia (Rachycentron canadum) in the Gulf of Thailand and Andaman Sea, in order to provide baseline information on the current population status and to assist in the design of sustainable cobia population management and aqua-farming policy development.
Cobia is a coastal pelagic fish which is pantropical in distribution, with the exception of the Pacific coast of the Americas (Shaffer and Nakamura 1989). It is a fast growing fish which is important for commercial and recreational fishing and has a high potential for aquaculture development (Benetti et al. 2008). Based on the FAO Fishery Statistics 2006, China and Taiwan are the main producing countries of cobia, but the production of cobia has also been reported in the Bahamas, Belize, the Dominican Republic, Mexico, Philippines, Puerto Rico, United States of America, and Vietnam (FAO 2012). In Thailand, there are currently many small cage farmers who capture juvenile cobia and raise them for a few months in river basins and estuaries. Small-scale production of cobia fingerling was started in early 2006 by Phuket and Rayong Coastal Fisheries Research and Development Centre (CFRDC). Increasing interest in cobia culture has led to projects aimed at developing hatchery technology for expanding cobia culture and increasing agro-economic development in coastal communities. The increasing promotion of cobia aquaculture will lead to a rise in stocking of increasingly domesticated strains of cobia in aquaculture operations. The practices of capturing wild fish to stock into cages will inevitably be replaced by domesticated stocks, increasing the potential influence of breeding programs on wild populations.
Cobia are multiple spawners (Brown-Peterson et al. 2001), with spawning of cobia occurring in both near shore and offshore waters with cobia forming aggregations, females releasing several hundred thousand to several million eggs and males releasing clouds of sperm (Shaffer and Nakamura 1989). Pre-juvenile cobia can be found in both near shore and offshore waters, whereas larger juveniles are mostly found near shore (Dawson 1971). Cobia display a variety of behaviors from association with small territories such as reefs or other structures to migrating to vast distances on currents or hitchhiking on large migratory marine animals such as whale sharks, sea turtles, whales, sharks, or rays (Shaffer and Nakamura 1989). A tagging study in the Gulf of Mexico showed a seasonal, long distance, migration of cobia from the Florida Keys to the Northern Gulf of Mexico, leaving the Florida Keys in spring and returning in the fall (Franks et al. 1991). Another tagging study in South Carolina, on the western edge of the Atlantic, showed that cobia move northward and southward along the coast, with few occasions moving into the Gulf of Mexico (Hammond 2001). Despite their relatively ubiquitous presence in the tropical seas of the world, little is known about the population genetic structure of this fish. Using mtDNA RFLP and cytochrome b sequence variation, it was suggested that individuals in the Gulf of Mexico, and along the south-eastern Atlantic coast from Virginia to Florida, are freely intermixed and can be considered as a single population (Hrincevich 1993). However, a recent study based on microsatellite markers showed significantly differentiated populations of cobia in the Persian Gulf and Oman Sea (Salari Aliabadi et al. 2008). While most of the recent research on cobia has aimed to improve the efficiency of cobia culture (Benetti et al. 2008; Fraser and Davies 2009), less effort has been dedicated to investigating the genetic diversity and structure of wild cobia populations in other geographic locations.
This study employs microsatellite markers to investigate genetic variation and population differentiation of wild cobia in the Gulf of Thailand and the Andaman Sea, which are geographically separated by an approximately 1,640-km stretch of the Thai–Malay Peninsula. Knowing the genetic structure of these populations can help to inform policy development and improve their effectiveness in controlling changes in population structure resulting from the introductions of domesticated fishes into the wild populations. This is particularly important for species such as cobia which are typically grown in cages either near shore or offshore in large-scale operations (Benetti et al. 2006; Miao et al. 2009) where potential escape events may occur. In addition, the narrow stretch of the Thai–Malay peninsula, with a narrowest width of approximately 56 km, also provides feasible translocation of marine stocks between the Andaman Sea and the Gulf of Thailand. Inter-ocean translocations of a total of 750 and 3,390 fingerlings from the Phuket CFRDC to fish cages along the Gulf of Thailand were conducted in 2006 and 2007, respectively. Cobia broodstock from the Phuket area (Andaman coast) were reportedly used for an experimental hatchery project at Rayong CFRDC (the Gulf of Thailand coast) in 2006. The production from this broodstock was distributed to local farmers along the Rayong coastline in the following year (2007). Fortunately, the samples analyzed in this study were collected before significant production from these facilities had begun. The study therefore provides baseline information of cobia natural populations prior to any contamination from translocated or hatchery-produced stocks.
Methods
Sample collection and DNA extraction
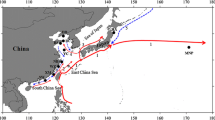
Fin clips from 268 cobia samples were collected from 6 study sites along coastal zones of the Gulf of Thailand and Andaman Sea with the assistance of the local fishing communities (Fig. 1; Table 1). The collecting sites in the Gulf of Thailand were in Trat (TR), Chonburi (CHN), and Prachuap Khiri Khan (PKK) provinces, whereas those in the Andaman Sea were in Phuket (PHU), Phangnga (PNG), and Ranong (RN) provinces. Samples were collected during June 2006–March 2007. All the fin samples were preserved in 100 % acetone. To extract genomic DNA, the fin tissue was homogenized in TNES buffer (10 mM Tris, pH 7.5, 400 mM NaCl, 100 mM EDTA, 0.6 % SDS). The gDNA was chloroform-extracted and precipitated in 100 % isopropanol. The DNA pellet was then washed in 70 % ethanol, air-dried, and resuspended in 0.1 × TE solution.
PCR amplification and allele scoring
Cobia microsatellite markers have been previously developed by Pruett et al. (2005). Of those, the nine most polymorphic loci, namely Rca 1-A11 (A11), Rca 1-B12 (B12), Rca 1-C04 (C4), Rca 1b-E02 (E2), Rca 1-E06 (E6), Rca 1-E11 (E11), Rca 1B-F06 (F6), Rca 1-G05 (G5), and Rca 1B-H09 (H9), were used in this study. The PCRs were performed in 20 μl volume of 50 mM Tris pH 9.2, 16 mM (NH4)2SO4, 2.25 mM MgCl2, 2 % DMSO, 0.1 % Tween-20, 0.2 mM dNTP3, 10 pmol of each forward and reverse primer, 1U GoTaq® Flexi DNA Polymerase (Promega), and the gDNA template in an amount that varied depending on which locus was being amplified. Following heat denaturation (94 °C 2 min), the reactions were carried out in 30 cycles of 94 °C for 30 s, optimal annealing temperature (dependent upon the locus being amplified) for 30 s, and 68 °C for 1 min, followed by 68 °C for 10 min. The optimized annealing temperature for A11 and F6 was 60 °C, for B12 and C4 was 58 °C, for E2 and H9 was 56 °C, for E6 and E11 was 53 °C, and for G5 was 48 °C. The optimized amount of gDNA template for A11, B12, E2, and F6 was 0.2 μg, for C4 and H9 was 0.4 μg, for E11 and G5 was 0.6 μg, and for E6 was 0.8 μg.
The PCR products were mixed with equal volumes of the 150 bp internal marker, which is a DNA fragment amplified from pGEM-T Easy vector (Promega), using primers 5′-ATACCTGTCCGCCTTTCTCC-3′ and 5′-TACCGGATAAGGCGCAG-3′. The denaturing buffer (10 mM NaOH, 0.05 % bromophenol blue, 20 mM EDTA in formamide) was then added to the mixed PCR products in a 1:1 ratio. The gel electrophoresis was run at constant 150 V for 6–8 h at 70 °C on 6 % denaturing 7 M urea, 0.5× Tris–Borate–EDTA polyacrylamide gels. The gel was silver-stained following the protocol described by Creste et al. (2001). Briefly, the gel was fixed in 10 % ethanol with 1 % acetic acid for 10 min, pre-treated in 1.5 % nitric acid for 3 min, and impregnated in 0.2 % AgNO3 for 20 min. In each step, the gel was washed with deionized water for 1 min, except that, after the impregnation step, the gel was washed with deionized water for 30 s, twice. The DNA bands were developed using cold (12 °C) developing solution (30 g/l Na2CO3, 0.54 ml 37 % formaldehyde), and the developing reaction was stopped by incubating the gel in 5 % acetic acid for 5 min. The gel images were captured using GeneSnap (Syngene). The low molecular weight DNA ladder (New England BioLabs) was included after every 8–10 lanes of samples. For each microsatellite locus, different alleles were called based on their relative sizes. The representative alleles of varying lengths were obtained after analyzing the first set of samples (at least 40 samples). The samples carrying these alleles were included in subsequent gels so that their alleles can be used as reference alleles. In the case that new alleles were found, these were also included in the subsequent gels. The samples which did not appear on the gels in positions close to their reference alleles were rerun in order to confirm the accuracy of the allele calling. The length and the number of base pairs of each allele reported in this study were estimated based on the results from GeneTools (Syngene) and on the information on the allele sizes and types of the repeat motif, previously reported by Pruett et al. (2005). In GeneTools, molecular weights were manually assigned to individual peaks in the low molecular weight DNA ladder tracks. To calculate molecular weights of the samples, the calibration curve was set to a log linear fit. The distance of the peaks was measured from the start line of the tracks, and the standard track nearest to the track being measured was used.
Descriptive genetic data
The number of alleles, the number of private alleles, and the observed and expected heterozygosity for each microsatellite locus were investigated using GDA (Lewis and Zaykin 2001). Allelic richness and genetic diversity were estimated using FSTAT (Goudet 2001). Significant differences in the levels of allelic richness and genetic diversity between the Gulf of Thailand and the Andaman Sea population groups were tested by 10,000 permutations using the same program. The inbreeding coefficients within populations (F is ) were used to test for deviations from Hardy–Weinberg equilibrium (HWE). Significant deviations from zero of the F is estimates were tested by the Markov chain method under 10,000 dememorization steps, 20 batches, and 5,000 iterations per batch, using GENEPOP (Rousset 2007). Genotypic disequilibrium between each locus pair across all samples was tested by the Markov chain method under 10,000 dememorization steps, 100 batches, and 5,000 iterations per batch, using the same program. Probability values from multiple tests were corrected based on Benjamini and Hochberg (1995) false discovery rate. The existence of null alleles was tested using MICRO-CHECKER (van Oosterhout et al. 2004). DETSEL (Vitalis et al. 2003) was used to test the neutrality of each microsatellite locus, in which μ was set at 0.0001, Ne was set at 5,000, and T 0 was set at 0.01. The distribution of F i and F j is generated by performing 10,000 coalescent simulations.
As a test for the source of apparent heterozygote deficiencies (inbreeding versus null alleles) and based on the argument that an inbred population would include individuals with differing degrees of inbreeding, we developed a test to see if homozygosity at different loci was correlated between individuals. Thus, in a population in linkage equilibrium, individual homozygosity at different loci should be uncorrelated if there is no inbreeding, even if apparent homozygosity is being created through the existence of null alleles. A positive correlation between loci in individual homozygosity will increase the variance in individual homozygosity. For each population, simulated samples were created in which the observed numbers of homozygotes and heterozygotes at all loci were permutated (by sampling without replacement across all the loci), and the variance in individual homozygosity of each simulated sample was compared to the observed variance in individual homozygosity in the sample. The proportion of simulated samples which had a variance that was greater than observed was used as a P value in a one-tailed statistical test. This was done with all loci except E6 and using only those individuals from the sample which had no missing genotypes at any of the loci.
Population differentiation analyses
The F ST parameter for each pair of populations was calculated according to Weir and Cockerham’s equation (1984), using GENEPOP (Rousset 2007). The statistical power of the dataset to detect significant population differentiation was investigated using POWSIM (Ryman and Palm 2006). The simulations with 1,000 runs were based on the number of samples with a complete multi-locus genotype, for either 7 loci or 6 loci (Table 2), using the various combinations of Ne (2,000, 5,000, 10,000) and t (20, 50, 100) that led to F ST of 0.0000, 0.0025, and 0.0050. Population differentiation was tested based on 210,000 randomizations of multi-locus genotypes between the sample pair using the log-likelihood G statistic (Goudet et al. 1996). The pairwise unbiased R ST from averaging variance components was estimated and tested for significant differentiation by 10,000 permutations, using RST CALC (Goodman 1997). The pairwise harmonic mean of D est across loci was calculated using SMOGD (Crawford 2010). To infer phylogenetic relationship between population samples, an unrooted neighbor-joining tree with 200 bootstrap replications was constructed based on the δμ2 genetic distance (Goldstein et al. 1995), using POPULATIONS (Langella 1999). The tree was plotted using TREEVIEWX (Page 1996). The population differentiation between the Gulf of Thailand and the Andaman Sea population groups was inferred by the analysis of molecular variance (AMOVA) and tested by 100,000 permutations, using ARLEQUIN (Excoffier et al. 1992). The level of missing genotype allowance was set at 10 %. Population isolation by distance was tested using a Mantel test (1967), in which the pairwise genetic isolation was measured by Slatkin’s linearized F ST (1995), and geographic distance was expressed by logarithms (log) of kilometers. The geographic distance between locations was determined by the shortest possible route for migration, which was roughly measured by the shortest straight coastline between locations. The test was conducted based on 100,000 permutations, using the same program.
STRUCTURE (Pritchard et al. 2000) was used to infer the population structure. This program searches for evidence of population structure based on the alleles possessed at multiple loci by each of the individuals in the total sample, without using any information about the geographic locations of the sampled individuals. In this program, the admixture model was used, and the Dirichlet parameter (α) for degree of admixture was set at 10. The model based on the assumption of correlated allele frequencies between populations was chosen in order to increase the sensitivity of the program to detect population structure differentiation (Falush et al. 2003). The potential numbers of population clusters (K) were explored ranging from 1 to 6. For each value of K, 20 runs of a Markov Chain Monte Carlo simulation with 100,000 burn-in iterations followed by a further 1,000,000 iterations were performed. The ad hoc statistic (ΔK) based on the rate of change in the log probability of data between successive K values was used to detect the uppermost hierarchical level of structure (Evanno et al. 2005).
Effective population size and gene flow
The population mutation rate parameter θ (θ = 4Neμ, where Ne is the effective population size and μ is the mutation rate per site) and the migration rate, M (M = m/μ, where m is the immigration rate per generation) were estimated by a maximum likelihood method, using a coalescent approach in MIGRATE-N (Beerli 2008). Three replications of estimations were conducted for the Gulf of Thailand and Andaman Sea population groups. In this program, the Brownian mutation model was used. The relative mutation rate was estimated from the data. The F ST estimates and UPGMA tree were used as starting parameters. The maximum likelihood was run for 10 short and 3 long chains with 10,000 and 100,000 recorded genealogies, respectively, from which the first 10,000 genealogies were discarded. One of every 20 reconstructed genealogies was sampled for both the short and long chains. The static heating scheme with 4 chains and temperatures of 1.0, 1.5, 3.0, and 10,000.0 was used. The model was chosen with both population groups having the same θ but allowing the migration rate (M) to be asymmetric.
Results
Microsatellite amplification success and tests for null alleles
The PCR amplification of all nine microsatellite loci was successfully optimized. However, there were problems with E2 and G5, when the amplifications were conducted on a number of samples. Both loci showed a low level of PCR success and a high number of homozygotes. This was more problematic for G5, where only 56 % of 91 samples were successfully amplified. The amplified products for G5 were found to be about 190–198 bp in length, which is smaller than reported in the study by Pruett et al. (2005) where they were said to be 274–282 bp. In addition, 89 % of the amplified samples for this locus were found to be homozygous. The high number of homozygotes and the difficulty of the PCR amplification of E2 and G5 suggest the possibility of a high frequency of null allele(s) at these two loci. Therefore, they were excluded from the analyses.
The other seven microsatellite loci were successfully amplified in all populations, although the extent of success varied between loci and population samples (Table 2). The average missing genotypes varied from 0.7 % (B12) to 20.5 % (E6). The sizes of the amplified products for each locus were consistent with those reported in the study by Pruett et al. (2005). The presence of null alleles was suggested in many loci and population samples (Table 2). Analyzed across samples, the presence of null alleles was suggested in all but one locus (H9). The signs of null alleles identified here are homozygote excesses that are seen for multiple homozygote classes. However, the loci that show a large deficiency in Ho relative to He are not the same loci that show the largest numbers of missing genotypes (Table 3).
Genetic diversity
The genetic diversity for each microsatellite locus is described in Table 3. The B12 locus showed the lowest gene diversity (0.515), whereas the F6 locus showed the highest gene diversity (0.954). The number of alleles of each locus ranged from 3 (B12) to 28 (F6) with an average of 17.7. The average allelic richness across loci and levels of gene diversity varied between populations from 11.9 to 13.1 and from 0.829 to 0.854, respectively. Comparing between the Gulf of Thailand and the Andaman Sea population groups, there were no significant differences in either the allelic richness (P = 0.710) or the gene diversity (P = 0.465). In addition, both population groups showed a low number of private alleles, of 1.4 on average.
Hardy–Weinberg equilibrium and linkage disequilibrium
Six of the seven loci studied (except for H9) showed significant overall heterozygote deficiencies (Table 3). On average, across loci, the observed heterozygosity was found to be significantly lower than expected based on HWE in all population samples (Table 3). Tests for genotypic disequilibrium between each locus pair across all samples showed a single significant linkage disequilibrium between C4 and H9. Because the physical location of each microsatellite locus studied is not known, it is not certain whether the significant linkage disequilibrium between C4 and H9 is due to physical linkage. Nevertheless, it is worth noting that population substructure can also create linkage disequilibrium. The observed and expected variances in homozygous loci per individual in each of the six populations are shown in Table 4. Also shown are the proportions of simulated samples, in which the homozygous loci were permuted across individuals, which exceed the observed variance. Here, one population, PNG, shows a significant upward deviation in its variance in homozygosity relative to expectation, even when the P value from the one-tailed test shown is doubled to create a two-tailed test. However, this significant result is not robust to correction for multiple testing.
Population differentiation and geographic distance correlation
The test for population differentiation is based on the assumption that the microsatellite makers are neutral. The tests by DETSEL showed no evidence of selection on any of the seven microsatellite loci studied since in all pairwise comparisons possible for the test, the observed F i and F j estimated from each of the loci all fell within the 95 % confidence region. As the populations deviated significantly from HWE, random mating cannot be assumed within these populations. The test for population differentiation was therefore performed based on genotypic data using the log-likelihood G statistic (Goudet et al. 1996). By this approach, only individuals with complete multi-locus genotypes were included in the test. The total number of samples with complete multi-locus genotypes is shown in Table 2. Because the E6 locus showed high percentages of missing genotypes in several populations, the population differentiation test was performed without the information from this locus, in order to increase the numbers of samples with complete multi-locus genotypes per population. Moreover, the results from POWSIM showed that the dataset with the exclusion of the E6 locus has a higher statistical power (66.1–67.4 % for F ST = 0.0025 and 98.1 % for F ST = 0.0050) to detect significant population differentiation than the dataset including all 7 loci, in which the number of samples demonstrating significant differentiation is lower (56.7–59.6 % for F ST = 0.0025 and 96.1–96.9 % for F ST = 0.0050). In both cases, the proportion of Type I error, when F ST was set at 0, varied from 4.1 to 7 %.
With the E6 locus excluded, the pairwise F ST values ranged from −0.0032 to 0.0081 (Table 5). The TR sample was found to be significantly differentiated from the CHN, PHU, PNG, and RN samples with P values, adjusted for multiple tests, less than 0.05. The pairwise R ST values ranged from −0.0109 to 0.0066. None of the R ST values were found to be significant. All population pairs show low values of the harmonic mean of D est, ranging from −0.0037 to 0.0312 (Table 6).
The phylogenetic relationship between cobia samples shows no obvious clustering between samples collected from the same geographic regions—the Gulf of Thailand and the Andaman Sea. The topology of the tree, in the Newick formulation, is [(PHU, PKK), RN], [CHN, (TR, PNG)]. Every node in the tree is supported by a bootstrap value lower than 35 %. The AMOVA analysis did not reveal significant genetic structure, either among sample populations or between the Gulf of Thailand and Andaman Sea sample groups (F CT = 0.0001, F SC = 0.001, F ST = 0.0011). Only 0.01 % of the genetic variation occurred between the two ocean basins, 0.1 % was due to differentiation among locations within each ocean basin and 99.89 % was due to variation within locations. STRUCTURE also did not detect any signs of population structure in the cobia samples. The log-likelihood and the ΔK are highest when K was set at 1 (Fig. 2). The summary Q plots showed all individuals being admixed and inferred to each population with the approximate proportion of 1/K for any K value (data not shown). The Mantel test revealed no significant correlation between genetic and geographic distance (R 2 = 0.0023, P = 0.328).
Effective population size and gene flow
The estimated population mutation parameter (θ = 4Neμ) and migration (M = m/μ) values, of cobia in the Gulf of Thailand and Andaman Sea estimated by MIGRATE-N, are shown in Table 7. The long-term effective population size (Ne) was obtained by assuming the average mutation rate of the microsatellites in cobia to be 1 × 10−4 mutation/generation/locus (Schug et al. 1997; Vigouroux et al. 2002; Whittaker et al. 2003; Yue et al. 2007). The number of immigrants per generation (Nm) was estimated by multiplying θ by M divided by 4. As can be seen in Table 7, the estimates varied between each run. The effective population size of the Gulf of Thailand population was estimated to be 5,785–8,465, and that of the Andaman Sea populations was estimated to be 5,093–12,268. In all three replications, cobia appear to migrate between the two ocean basins with fairly similar migration rates. However, the estimated effective population size and migration rate reported in this study must be taken with caution. This is because the estimation by MIGRATE-N is based on an assumption of random mating in the populations, which is apparently not the case for the microsatellite markers used in this study. In addition, the actual microsatellite mutation rate in cobia is not known.
Discussion
Genetic diversity and Hardy–Weinberg equilibrium
Analysis of the natural cobia population in the Gulf of Thailand and Andaman Sea has shown levels of gene diversity at 0.844 and 0.837, respectively. These levels are similar to what have been found in the cobia samples from the northern Gulf of Mexico (0.804, Pruett et al. 2005) and Iran (0.874, Salari Aliabadi et al. 2008). It should be mentioned that the value of gene diversity of the northern Gulf of Mexico samples shown here was estimated using the information from the same microsatellite loci as in this study. Most of the loci studied show a moderately high number of alleles with an average across loci of 17.7. These levels of gene diversity and allelic richness in cobia are similar to the averages found in other marine fishes (DeWoody and Avise 2000). When compared to the northern Gulf of Mexico and Iranian samples (Pruett et al. 2005; Salari Aliabadi et al. 2008), each locus studied showed a higher number of alleles. This could be simply because the sample sizes in our study are higher, and therefore, there is a higher chance of finding more alleles. Nevertheless, one exception is the B12 locus, where only 3 alleles were found in the Gulf of Thailand and Andaman Sea samples (n = 247) whereas as many as 7 alleles were found in the northern Gulf of Mexico samples (n = 24). If this is not because of the differences in the scoring methods, it is possible that the cobia populations in the northern Gulf of Mexico and in Thailand contain different genetic compositions.
All populations and all but one microsatellite locus studied (H9) were found to have significantly lower heterozygosity than HWE with positive F is (Table 3). When the analyses were carried out with the data excluding all missing genotypes, the values of F is , even though different from those shown in Table 3, did not show any consistent trends of bias. In addition, in the data without missing genotypes, the significant heterozygote deficiencies, with positive F is , were still observed in all populations and all but one of the microsatellite loci studied (H9), suggesting that the estimations of heterozygote deficiencies in Table 3 were not strongly influenced by the missing genotypes. In general, heterozygote deficiencies can be observed if (1) the locus is sex-linked, (2) there are null alleles, (3) the population has been inbreeding within its own group, or (4) there is population substructure (Wahlund effect). The possibility of any loci being sex-linked can be ruled out, as each of the loci studied was found to be heterozygous in both male and female samples. The other three possible explanations are, however, possible. Null alleles are non-amplified alleles that are caused by nucleotide variations in the primer binding sites. If one of the two alleles fails to be amplified, the sample would be scored as a homozygote instead of a heterozygote, causing an overall heterozygote deficiency. In our study, all but one locus (H9) show heterozygote deficiencies, and the samples showed an average proportion of missing genotypes across loci of 7.88 %, with the lowest percentage being 0.7 % (B12) and the highest percentage 20.5 % (E6). The questions are whether null alleles are truly present at all these microsatellite loci and whether the frequencies of these null alleles are so high that individuals homozygous for null alleles are what are being identified as the missing genotypes. However, while the PCR amplification from samples homozygous for null alleles would fail and the genotype of these samples could not be scored, missing genotypes do not necessarily indicate that the individual is homozygous for null alleles. This is because a failure of PCR amplification can be caused by factors other than null alleles. For example, there could be DNA template degradation and/or impurity. In the presence of PCR inhibitors, a low primer site fidelity and a low level of interference can result in the PCR failure, the extent of which can vary between primer pairs. However, for the B12 and C4 loci, the possibility of null alleles were suggested in the TR and the PKK populations, respectively, and here, there are no missing genotypes (Table 2). This showed that even when the amplification should potentially work, these samples were still showing too few heterozygotes. This evidence suggests that the heterozygote deficiencies in these two loci are due to the presence of null alleles. Whether the null alleles are also present in the other four loci that show significant heterozygote deficiencies is not clear, but the high percentages of missing genotypes, particularly F6, E6 and E11, cannot be explained by all these individuals being homozygous for null alleles. If these missing genotypes at these loci were all null allele homozygotes, then the frequencies of missing genotypes would (from the Hardy–Weinberg formula) suggest null allele frequencies of, respectively, 31, 45, and 27 % at these three loci. Null allele frequencies this large would create much larger F is estimates than are seen at any of these loci (Table 3). Thus, the evidence suggests that the failure to amplify must be due to reasons other than null alleles, and thus null alleles are not necessarily the explanation for the heterozygote deficiencies in every locus studied. It is interesting that heterozygote deficiency was also observed in the Persian Gulf and Oman Sea cobia populations at most of the microsatellite loci studied—A4, A10, E2, E8A, F6, F7, and H9—(Salari Aliabadi et al. 2008). Given that it is unlikely that we would observe, independently, null alleles at almost all loci studied, the presence of significant heterozygote deficiencies in most of the loci studied could be a result of non-random mating. Cobia has been observed in the wild to form small spawning aggregate groups which then separate into groups of two or more, before releasing eggs and sperm (Shaffer and Nakamura 1989). In addition, cobia often associate with small territories, where high concentrations of cobia can be found (Franks et al. 1991; our observations) and seasonally migrate over large distance to avoid cold water (Franks et al. 1991), which leads to the hypothesis that local populations in the areas where there is no seasonal impetus for migration such as Thailand may be substructured (a Wahlund effect), containing genetically differentiated resident and transient groups. However, the inability of STRUCTURE to distinguish between resident and transient populations suggests a relatively constant admixture of the two populations throughout the study area rather than the existence of separate populations. It is possible that the samples collected in each study site may contain some extent of inbreeding while also experiencing admixture between the resident and transient groups, with some components of these admixed local populations regularly making a long distance migrations between ocean basins, causing population homogeneity on a larger geographic scale. In order to differentiate between inbreeding and null alleles as the cause of the apparent heterozygote deficiencies, we argue, following Overall and Nichols (2005) that, if there is inbreeding, there will be variation between individuals in the degree of their inbreeding. This will create a correlation between loci in individual homozygosity, as an individual homozygous at one locus will have a higher chance of being inbred and thus a higher chance of being homozygous at other loci. The consequence is that inbreeding will be manifest as a greater than expected variance in individual homozygosity (summed across loci). We tested for this effect with simulated samples in which homozygotes at the different loci were permuted (Table 4). While one population (PNG) showed a significant excess in variance in homozygosity, this could potentially be a Type I error arising from multiple testing, so it is still not clear whether the datasets have been affected by inbreeding. An argument is that the population homogeneity observed could be because the samples studied were collected during the non-reproductive season; the studied samples could then be a mixed group of migrating individuals from differentiated populations, which could create a pattern of genetic homogeneity, blurring the true population structure. The spawning season of cobia in Thailand is currently not known. Whether or not the spawning can occur all year round or whether the spawning season in Andaman Sea and the Gulf of Thailand is the same and whether cobia in Thai waters show a seasonal migration and site fidelity still require further investigations. As can be seen in Table 1, most of the samples were adults, which were collected in the months of October–November, but juveniles were collected from PKK (the Gulf of Thailand) in October and from PHU (Andaman Sea) in June. It is possible that cobia in the two ocean basins may have different spawning seasons, and most of the samples used in the study were collected just a month or two after the spawning season in the Gulf of Thailand. However, since the majority of the samples were collected from both the Andaman Sea and the Gulf of Thailand at approximately the same time, it implies that cobia may not show a strong seasonal migration in the same way as they do in the Atlantic and the Gulf of Mexico. In the case of Atlantic cobia, it will be very difficult to collect abundant samples from South Carolina and Florida at the same period of time during non-reproductive season. Therefore, the effect of seasonal migration on the population homogeneity of Thai cobia is still questionable. More studies on life history of cobia in these areas are needed. Whether or not there are resident and transient populations in these areas, and to what extent these groups interbreed requires further investigation with more genetic markers, coupled with tagging studies.
Tests on population differentiation based on geographic locations
Southeast Asia is the most obvious geographic boundary between the Pacific and the Indian Oceans. The Thai–Malay peninsula separates the Andaman Sea (which is part of the Indian Ocean) and the Gulf of Thailand (which is part of the South China Sea). Interestingly, wild cobia population samples show no marked population structure, even over the long barrier of the Thai–Malay peninsula. Most pairwise population comparisons show low and non-significant values of F ST and R ST (Table 5), whether evaluated within the same ocean basin or between the Gulf of Thailand and the Andaman Sea. In principle, the population differentiation test relies heavily on the statistical power of the dataset, which is a function of the level of variation at the loci used, the sample size, and levels of genetic distance between populations. By simulation analyses, the data excluding the E6 locus show a moderate statistical power (66.1–67.4 %) to detect population differentiation if the F ST = 0.0025, but a very high statistical power (98.1 %) if the F ST = 0.0050. Although the confidence of pairwise comparisons where F ST < 0.0050 could be improved with the evaluation of more loci and larger sample sizes, the non-significant F ST between population pairs presented here is concordant with the apparent lack of population structure as evaluated through the AMOVA test. Significant pairwise F ST values were detected most often between the TR sample and the others. However, the significance levels are low (corrected P values ≤0.05—Table 5). When combined with the lack of significant pairwise R ST and the lack of correlation between genetic and geographic distance, the observed significant pairwise F ST between the TR sample and the others may not reflect true population differentiation of the species in this area. In addition, it has been demonstrated that the presence of null alleles can cause an overestimation of genetic difference (Chapuis and Estoup 2007). It is possible that the low but significant levels of genetic divergence between different populations observed could be due to the effect of null alleles, since the analyses here were conducted without corrections for null alleles for any of the loci.
The fact that this study shows no significant population structure or isolation by distance of cobia populations along the Thai–Malay peninsular coast does not rule out the possibility of significant differentiation between cobia populations that are more distantly distributed. Compared to Pruett and colleagues’ study (2005), some of the microsatellite alleles found in the Gulf of Mexico samples are not observed in the Gulf of Thailand and Andaman Sea populations. Considering that cobia have demonstrated the ability to migrate to distances over 660 miles or 1,062 km (Hammond 2001), it would be interesting to see at what geographic distances the populations start to show isolation by distance. The pattern of cobia population structure in the Persian Gulf and Oman Sea was found to be different from that observed in the Gulf of Thailand and Andaman samples. The populations in the Persian Gulf were found to be significantly differentiated from each other and from those in the Oman Sea (Salari Aliabadi et al. 2008). However, the significance levels in their study are low (P values ≤0.05), and were not corrected for multiple tests. The difference in the pattern of population structure between the Thai and Iranian cobia, if it exists, could be due to the differences in the geographic and oceanographic features and/or the evolutionary history of the studied areas.
In general, the high gene flow between populations can be explained either by adult individuals migrating over large distances to interbreed with distant groups or by adult individuals breeding with near neighbor groups and their larvae and juveniles drifting to distant areas under the influence of ocean currents. In the case of cobia in the Gulf and Andaman Sea, both explanations may account for its population homogeneity. Cobia is well known for hitchhiking on large marine animals and migrating to long distances on its own (Shaffer and Nakamura 1989). Population genetic studies on whale sharks, a frequently observed hitchhiking host of cobia, have shown that whale shark migrations can cover vast distances (as much as 13,000 km) and that their population shows no marked structure between the Indian and Pacific Oceans (Castro et al. 2007; Schmidt et al. 2009). This behavior of cobia therefore has the potential to increase its migration range to include geographic regions separated by vast distances. The rates of cobia migration between the Gulf of Thailand and Andaman Sea were estimated to be roughly equivalent (Table 7). Oceanographic data on currents in the region suggest a possible mechanism for larval and juvenile fish from the Gulf of Thailand populations to migrate to the Andaman Sea during the northeast monsoon season (Wyrtki 1961; Yanagi et al. 2001; Pang and Tkalich 2003) and from the Andaman Sea populations to the Gulf of Thailand, during the southwest monsoon season (Brown 2007; Fairbridge 1966; Wyrtki 1961).
Status of cobia population and implication for fisheries management
The cobia population in the Gulf of Thailand and Andaman Sea shows a moderate to high level of genetic diversity. At present, there is no regular collection of data to show changes in the level of genetic diversity of cobia over time. Additionally, it has been shown that a high or unchanged level of genetic diversity may not be able to fully reflect the status of population exploitation if the effective population size of the species is large (Hoarau et al. 2005). However, due to the small and undervalued domestic market for cobia, it is possible that cobia fisheries in Thailand may not yet be overexploited. The situation of cobia fisheries may change in the future during the initial stages of cobia culture development, as cobia culture gains popularity and the export and domestic markets strengthen, or as alternative species are sought to replace the depleted resources of traditional fisheries. Fisheries and marine resource policies to protect cobia from overexploitation should be in place prior to increases in pressure on wild stocks. One of the greatest pressures on cobia in Thailand today may be commercial trawling for demersal fish and shrimp. We observed incidents in which a large number of cobia in juvenile size classes were captured by this practice.
The estimates of the long-term effective population size of cobia in this study, although uncertain, are low compared to that of gray whale (Alter et al. 2007). It should be mentioned that the estimation of Ne using this coalescent method depends largely on the mutation rate at the markers used. Since the mutation rates of cobia microsatellite markers are not known, the true Ne values could differ from those in Table 7. Moreover, the estimated Ne here is a long-term average, which could be very different from its current value—the value that is most important for conservation. It is possible that the current effective population size of cobia is much larger than in the past, and the long-term effective population size (which determines the level of variation now observed), since it is the harmonic mean of the effective sizes at different times, is very much affected by those time intervals when the population size was small. The evidence that fishermen can catch many cobia in the wild suggests that the current population size of cobia is not small.
The homogeneity of cobia stocks sampled in this study suggests that the population investigated may range across no less than 8 countries (Myanmar, India, Thailand, Malaysia, Indonesia, Singapore, Cambodia, and Vietnam) and may likely include more. Fisheries management should therefore be considered at both national and international levels to ensure the preservation of genetic diversity and the sustainability of local and regional fisheries. It was found that populations appearing homogeneous and panmictic in neutral microsatellite markers may possess locally adapted populations (Hauser and Ward 1998). Cobia show a high degree of site fidelity (Hammond 2001), suggesting a possible cue which brings subsequent generations back to the same seasonal spawning grounds from which they originated. This could have implications for adaptation to specific sites or fitness reductions in individuals returning to a specific location if genes from fish originating from specific sites are diluted or replaced by those from domesticated or translocated groups over time. The development of fisheries management policy regarding intentional translocation of stocks from alternate ocean basins should be guided by the precautionary principle, and efforts should be made to reduce the instance of artificial translocations. The prospect of large-scale cage culture of cobia in the Gulf of Thailand using fingerlings or broodstock originating from the Andaman Sea may create the potential to unintentionally alter the population structure of native Gulf of Thailand fishes. More microsatellite loci and a larger sample size could be used to increase the power of analyses of fine scale differentiation, and mitochondrial DNA analysis can be used to reach further back into the potential cladistic origins of each group. There are many questions that remain about phenotypic variation and potential local adaptation, which need to be answered to inform policy development in fisheries and aquaculture development. Tagging and tracking studies, with integrated genetic sampling, should be conducted to provide more detail about the nature, pattern, and route of migration, and the temporal and spatial location of cobia spawning grounds.
References
Alter SE, Rynes E, Palumbi SR (2007) DNA evidence for historic population size and past ecosystem impacts of gray whales. PNAS 104:15162–15167
Beerli P (2008) MIGRATE-n estimation of population sizes and gene flow using the coalescent. Available from http://popgen.scs.fsu.edu/Migrate-n.html
Bekkevold D, Hansen MM, Nielsen EE (2006) Genetic impact of gadoid culture on wild fish populations: predictions, lessons from salmonids, and possibilities for minimizing adverse effects. ICES J Mar Sci 63:198–208
Benetti D, Brand L, Collins J et al (2006) Can offshore aquaculture of carnivorous fish be sustainable? Case studies from the Caribbean. World Aquac Mag 37:44–47
Benetti DD, Orhun MR, Sardenberg B et al (2008) Advances in hatchery and grow-out technology of cobia Rachycentron canadum (Linnaeus). Aquac Res 39:701–711
Benjamini Y, Hochberg T (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc B 85:289–300
Brown BE (2007) Coral reefs of the Andaman Sea—an integrated perspective. In: Gibson RN, Atkinson RJA, Gordon JDM (eds) Oceanography and marine biology: an annual review, vol 45. CRC Press, Boca Raton, pp 173–194
Brown-Peterson NJ, Overstreet RM, Lotz JM et al (2001) Reproductive biology of cobia, Rachycentron canadum, from coastal waters of the southern United States. Fish Bull 99:15–28
Castro ALF, Stewart BS, Wilson SG et al (2007) Population genetic structure of earth’s largest fish, the whale shark (Rhinocodon typus). Mol Ecol 16:5183–5192
Chapuis M, Estoup A (2007) Microsatellite null alleles and estimation of population differentiation. Mol Biol Evol 24:621–631
Crawford NG (2010) SMOGD: software for the measurement of genetic diversity. Mol Ecol Res 10:556–557
Creste S, Tulmann Neto A, Figueira A (2001) Detection of single sequence repeat polymorphisms in denaturing polyacrylamide sequencing gels by silver staining. Plant Mol Biol Rep 19:299–306
Dawson CE (1971) Occurrence and description of prejuvenile and early juvenile Gulf of Mexico cobia, Rachycentron canadum. Copeia 1971:65–71
DeWoody JA, Avise JC (2000) Microsatellite variation in marine, freshwater and anadromous fishes compared with other animals. J Fish Biol 56:461–473
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Fairbridge RW (1966) The encyclopedia of oceanography. Reinhold Publishing Corporation, New York
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
FAO (2012) Cultured aquatic species information programme Rachycentron canadum (Linneaeus, 1766). Available from http://www.fao.org/fishery/culturedspecies/Rachycentron_canadum/en
Fjalestad KT, Gjedrem T, Gjerde B (1993) Genetic improvement of disease resistance in fish: an overview. Aquaculture 111:65–74
Fleming IA, Hindar K, Mjølnerød IB et al (2000) Lifetime success and interactions of farm salmon invading a native population. Proc R Soc Lond B 267:1517–1523
Franks JS, Zuber MH, McIlwain TD (1991) Trends in seasonal movements of cobia, Rachycentron canadum, tagged and released in the northern Gulf of Mexico. J Miss Acad Sci 36:55
Fraser TWK, Davies SJ (2009) Nutritional requirements of cobia, Rachycentron canadum (Linnaeus): a review. Aquac Res 40:1219–1234
Gjøen HM, Bentsen HB (1997) Past, present, and future of genetic improvement in salmon aquaculture. ICES J Mar Sci 54:1009–1014
Goldstein DB, Linares AR, Cavalli-Sforza LL et al (1995) An evaluation of genetic distances for use with microsatellite loci. Genetics 139:463–471
Goodman SJ (1997) RST calc: a collection of computer programs for calculating estimates of genetic differentiation from microsatellite data and determining their significance. Mol Ecol 6:881–885
Goudet J (2001) FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3). Available from http://www2.unil.ch/popgen/softwares/fstat.htm
Goudet J, Raymond M, de Meeüs T et al (1996) Testing differentiation in diploid populations. Genetics 144:1933–1940
Hammond DL (2001) Status of the South Carolina fisheries for cobia. South Carolina Department of Natural Resources Technical Report Number 89
Hauser L, Ward RD (1998) Population identification in pelagic fish: the limits of molecular markers. In: Carvalho GR (ed) Advances in molecular ecology. NATO Science Series: life sciences, vol 36. IOS Press, Amsterdam, pp 191–224
Hauser L, Adcock GJ, Smith PJ et al (2002) Loss of microsatellite diversity and low effective population size in an overexploited population of New Zealand snapper (Pagrus auratus). PNAS 99:11742–11747
Hena A, Kamal M, Mair GC (2005) Salinity tolerance in superior genotypes of tilapia, Oreochromis niloticus, Oreochromis mossambicus and their hybrids. Aquaculture 247:189–201
Hindar K, Fleming IA, McGinnity P et al (2006) Genetic and ecological effects of salmon farming on wild salmon: modelling from experimental results. ICES J Mar Sci 63:1234–1247
Hoarau G, Boon E, Jongma DN et al (2005) Low effective population size and evidence for inbreeding in an overexploited flatfish, plaice (Pleuronecter platessa L.). Proc R Soc Lond B 272:497–503
Hrincevich AW (1993) Analysis of cobia, Rachycentron canadum, population structure in the northern Gulf of Mexico using mitochondrial DNA. MSc thesis, University of Southern Mississippi
Hutchings JA, Fraser DJ (2008) The nature of fisheries- and farming-induced evolution. Mol Ecol 17:294–313
Hutchinson WF, van Oosterhout C, Rogers SI et al (2003) Temporal analysis of archived samples indicates marked genetic changes in declining North Sea cod (Gadus morhua). Proc R Soc Lond B 270:2125–2132
Kuparinen A, Merilä J (2007) Detecting and managing fisheries-induced evolution. TREE 22:652–659
Langella O (1999) Populations 1.2.30. Available from http://bioinformatics.org/~tryphon/populations/
Lewis PO, Zaykin D (2001) Genetic data analysis: computer program for the analysis of allelic data. Version 1.0 (d16c). Available from http://hydrodictyon.eeb.uconn.edu/people/plewis/software.php
Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220
McGinnity P, Prodöhl P, Ferguson K et al (2003) Fitness reduction and potential extinction of wild populations of Atlantic salmon, Salmo salar, as a result of interactions with escaped farm salmon. Proc R Soc Lond B 270:2443–2450
Miao S, Jen CC, Huang CT, Hu S (2009) Ecological and economic analysis for cobia Rachycentron canadum commercial cage culture in Taiwan. Aquac Int 17:125–141
Overall ADJ, Nichols RA (2005) A method for distinguishing consanguinity and population substructure using multilocus genotype data. Mol Biol Evol 18:2048–2056
Page RDM (1996) TreeView: an application to display phylogenetic trees on personal computers. Comp Appl Biosci 12:357–358
Palumbi SR (2003) Population genetics, demographic connectivity, and the design of marine reserves. Ecol Appl 13:S146–S158
Pang WC, Tkalich P (2003) Modeling tidal and monsoon driven currents in the Singapore Strait. Singap Marit Port J 2003:151–162
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Pruett CL, Saillant E, Renshaw M et al (2005) Microsatellite DNA markers for population genetic studies and parentage assignment in cobia, Rachycentron canadum. Mol Ecol Notes 5:84–86
Rezk MA, Smitherman RO, Williams JC et al (2003) Response to three generations of selection for increased body weight in channel catfish, Ictalurus punctatus, grown in earthen ponds. Aquaculture 228:69–79
Rousset F (2007) GENEPOP’007: a complete re-implementation of the GENEPOP software for Windows and Linux. Mol Ecol Resour 8:103–106
Ryman N, Palm S (2006) POWSIM: a computer program for assessing statistical power when testing for genetic differentiation. Mol Ecol Notes 6:600–602
Salari Aliabadi MA, Rezvani Gilkolaei S, Savari A et al (2008) Microsatellite polymorphism in Iranian populations of cobia (Rachycentron canadum G.). Biotechnology 7:775–780
Schmidt JV, Schmidt CL, Ozer F et al (2009) Low genetic differentiation across three major ocean populations for the whale shark, Rhincodon typus. PLoS One 4:e4988
Schug MD, Mackay TFC, Aquadro CF (1997) Low mutation rates of microsatellite loci in Drosophila melanogaster. Nat Genet 15:99–102
Shaffer R, Nakamura E (1989) Synopsis of biological data on the cobia, Rachycentron canadum (Piseces: Rachycentridae). NOAA Technical Report NMFS 82. FAO Fisheries Synopsis 153
Slatkin M (1995) A measure of population subdivision based on microsatellite allele frequencies. Genetics 139:457–462
Smith PJ, Francis R, McVeagh M (1991) Loss of genetic diversity due to fishing pressure. Fish Res 10:309–316
Turner TF, Wares JP, Gold JR (2002) Genetic effective size is three order of magnitude smaller than adults census size in an abundant, estuarine-dependant marine fish (Scianops ocellatus). Genetics 162:1329–1339
van Oosterhout C, Hutchinson WF, Wills DPM et al (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Vigouroux Y, Jaqueth JS, Matsuoka Y et al (2002) Rate and pattern of mutation at microsatellite loci in maize. Mol Biol Evol 19:1251–1260
Vitalis R, Dawson K, Boursot P et al (2003) DetSel 1.0: a computer program to detect markers responding to selection. J Hered 94:429–431
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Whittaker JC, Harbord RM, Boxall N, Mackay I, Dawson G, Sibly RM (2003) Likelihood-based estimation of microsatellite mutation rates. Genetics 164:781–787
Wyrtki K (1961) Physical oceanography of the Southeast Asian waters. NAGA report 2
Yanagi T, Sachoemar SI, Takao T et al (2001) Seasonal variation of stratification in the Gulf of Thailand. J Oceanogr 57:461–470
Yue G, David L, Orban L (2007) Mutation rate and pattern of microsatellites in common carp (Cyprinus carpio L.). Genetica 129:329–331
Acknowledgments
We would like to thank all the local fishermen and fishing communities for providing the cobia samples. We also would like to thank Phuket Coastal Fisheries Research and Development Centre for providing the samples for technique optimization and both Phuket and Rayong Coastal Fisheries Research and Development Centres for the information on cobia hatchery operations. This work is supported by the Office of the Higher Education Commission and Thailand Research Fund grant number MRG4980195.
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License (https://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Phinchongsakuldit, J., Chaipakdee, P., Collins, J.F. et al. Population genetics of cobia (Rachycentron canadum) in the Gulf of Thailand and Andaman Sea: fisheries management implications. Aquacult Int 21, 197–217 (2013). https://doi.org/10.1007/s10499-012-9545-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10499-012-9545-1